
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19455254-19455375 + | AAGTACAATA---TCTTGGT--------TGGCGGCACAATCTGCAATCTACGCCACTGGCTTACGTTGC-------AA-TCGAAAATCGTGTCCAGTGCCGTAAATTGCAGTTGTGTGAA--CGCAAACGGCAGCAG--------------------CGAAGG |
| droSim1 | chrX_random:5079001-5079113 + | AAGTA------------GAT--------TGGCGGCACAATCTGCAATCTACGCCACTGGCTTACGTTGT-------GG-TCGAGAATCGTGTCCAGTGCCGTATATTGCAGTTGTGTGGA--CGCAAACGGCAGCAG--------------------CGAAGG |
| droSec1 | super_8:1737859-1737971 + | AAGTA------------GAT--------TGGCGGCACAATCTGCAATCTACGCCACTGGCTTACGTTGT-------AG-TCGAGAATCGTGTCCAGTCCCGTAAATTGCAGTTGTGTGGA--CGCAAACGGCAGCAG--------------------CGAAGG |
| droYak2 | chrX:11883747-11883862 - | AAGTGCATCC------AGGT--------GGGCGGCACAGCCAGCACTCTACGCCGCTGGCTTACGTTGT-------AC-TCGGAAATCGTGTCCAGTGCCGTTGATTGCAGTTGTGTGGA--CGCAAAGGGCAGCG-----------------------GAGA |
| droEre2 | scaffold_4690:9691187-9691301 + | AAGTGCATCC-------AGT--------GGGCGGCACAACCAGCATTCTACGCCGCTGGCTTACGTTGT-------AC-TCGAAAATCGTGTCCAGTGCCGTTGATTGCAATTGTGTGGA--CGCAAAGGGCAGCG-----------------------GAGG |
| droAna3 | scaffold_13417:2460507-2460635 - | GAGTGCGGACAGGGCAAGCT--------CGGCGGCACAGACAGCATTCGACGCTGCTGGCTTACGTGGT-------GG-TTGATATCCGTGTCCAGTGACGTTGATTGCAGTTGTGTAGA--CGCGAATTACAGCGACCG------A----------TAGTTG |
| dp4 | Unknown | ------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 | super_14:1991206-1991216 - | AACAAC--------------------------------------------------------------------------------------------------------------------------------------------------------AACTA |
| droWil1 | scaffold_181096:7961115-7961220 + | -------------------T--------TGGCAACACAGGCAGCATTCGACGACGCTGGCTTACGTTAAAAAC---AA-ACGATATCCGTGTCCAGTGACGTTGATTGCAGTTGTGTA------CAATGATTAGCGG--------------------CGATTT |
| droVir3 | scaffold_13042:902659-902773 - | ACA-----------------------------GGCACA-GCCGTACTCTACGCTTTTGGGA-ACGAG--CTCTTTGAC----GCACTCGGTTCCATTGCCGTTGAGTAGAGCTGTCCTGA--TGCATATCTCAACG---T------ATTGATTCAAACAAGAA |
| droMoj3 | scaffold_6328:2897048-2897148 - | CG------------------------------GGTACA-GCCGTACTCTACGCTTTTGGGA-ACGAG--CAATCATACAAAGCCACTCGGTTCCATAGCCGTTGAGTAGAGCTGCCCCCGTTTGCACATCACAAC---------------------------- |
| droGri2 | scaffold_15081:2040911-2041055 - | AAGTACGGCA---GTTCGATAACCAACAC--AGGTCCA-GCCGTATTCAGCGCTTTTGGAA-GTAAG--GAACATGAT----TCTCCTAGTTTCAATGCCGCTGAGTAAAGCTGTCCCAA--TGCAAATCGCAACG---TAACTGAATCGAATCAATCAATAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19455254-19455375 + | AAGTACAAT---ATCTTGGT--------TGGCGGCACAATCTGCAATCTACGCCACTGGCTTACGTTGC-------AA-TCGAAAATCGTGTCCAGTGCCGTAAATTGCAGTTGTGTGAA--CGCAAACGGCAGCAG--------------------CGAAGG |
| droSim1 | chrX_random:5079001-5079113 + | AAGTA------------GAT--------TGGCGGCACAATCTGCAATCTACGCCACTGGCTTACGTTGT-------GG-TCGAGAATCGTGTCCAGTGCCGTATATTGCAGTTGTGTGGA--CGCAAACGGCAGCAG--------------------CGAAGG |
| droSec1 | super_8:1737859-1737971 + | AAGTA------------GAT--------TGGCGGCACAATCTGCAATCTACGCCACTGGCTTACGTTGT-------AG-TCGAGAATCGTGTCCAGTCCCGTAAATTGCAGTTGTGTGGA--CGCAAACGGCAGCAG--------------------CGAAGG |
| droYak2 | chrX:11883747-11883862 - | AAGTGCATC------CAGGT--------GGGCGGCACAGCCAGCACTCTACGCCGCTGGCTTACGTTGT-------AC-TCGGAAATCGTGTCCAGTGCCGTTGATTGCAGTTGTGTGGA--CGCAAAGGGCAGCG-----------------------GAGA |
| droEre2 | scaffold_4690:9691187-9691301 + | AAGTGCATC-------CAGT--------GGGCGGCACAACCAGCATTCTACGCCGCTGGCTTACGTTGT-------AC-TCGAAAATCGTGTCCAGTGCCGTTGATTGCAATTGTGTGGA--CGCAAAGGGCAGCG-----------------------GAGG |
| droAna3 | scaffold_13417:2460507-2460635 - | GAGTGCGGACAGGGCAAGCT--------CGGCGGCACAGACAGCATTCGACGCTGCTGGCTTACGTGGT-------GG-TTGATATCCGTGTCCAGTGACGTTGATTGCAGTTGTGTAGA--CGCGAATTACAGCGACCG------A----------TAGTTG |
| droWil1 | scaffold_181096:7961115-7961220 + | -------------------T--------TGGCAACACAGGCAGCATTCGACGACGCTGGCTTACGTTAAAAAC---AA-ACGATATCCGTGTCCAGTGACGTTGATTGCAGTTGTGTA------CAATGATTAGCGG--------------------CGATTT |
| droVir3 | scaffold_13042:902659-902773 - | ACA-----------------------------GGCACA-GCCGTACTCTACGCTTTTGGGA-ACGAG--CTCTTTGAC----GCACTCGGTTCCATTGCCGTTGAGTAGAGCTGTCCTGA--TGCATATCTCAACG---T------ATTGATTCAAACAAGAA |
| droMoj3 | scaffold_6328:2897048-2897148 - | CG------------------------------GGTACA-GCCGTACTCTACGCTTTTGGGA-ACGAG--CAATCATACAAAGCCACTCGGTTCCATAGCCGTTGAGTAGAGCTGCCCCCGTTTGCACATCACAAC---------------------------- |
| droGri2 | scaffold_15081:2040911-2041055 - | AAGTACGGC---AGTTCGATAACCAACAC--AGGTCCA-GCCGTATTCAGCGCTTTTGGAA-GTAAG--GAACATGAT----TCTCCTAGTTTCAATGCCGCTGAGTAAAGCTGTCCCAA--TGCAAATCGCAACG---TAACTGAATCGAATCAATCAATAA |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
| dG=-27.7, p-value=0.009901 | dG=-27.4, p-value=0.009901 |
|---|---|
 |
 |
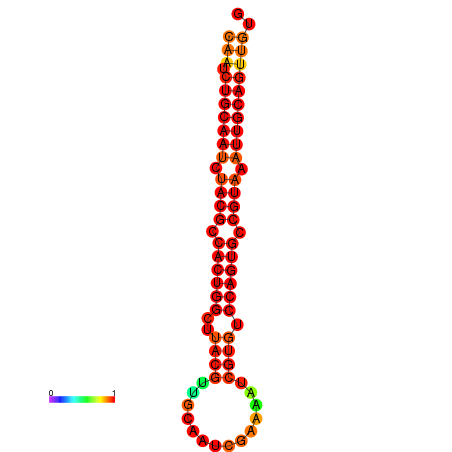
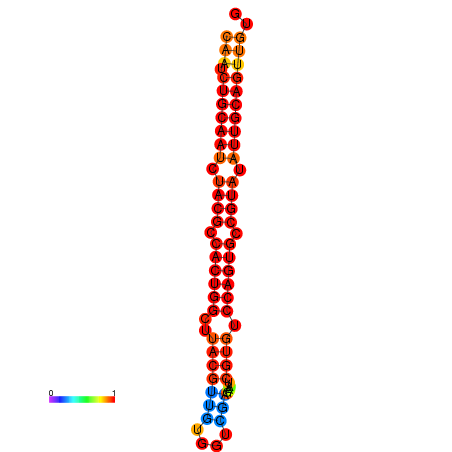
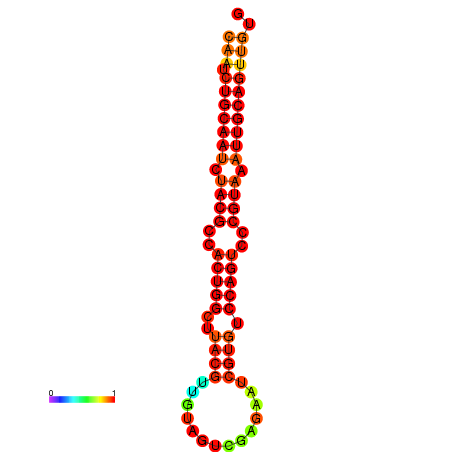
| dG=-27.7, p-value=0.009901 | dG=-27.6, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
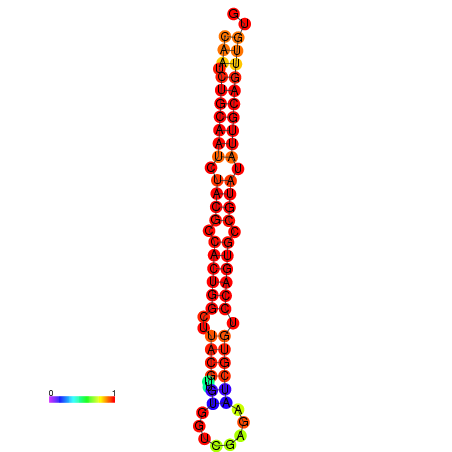 |
| dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|
 |
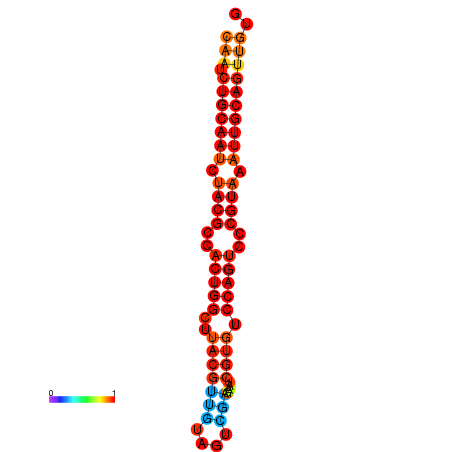 |
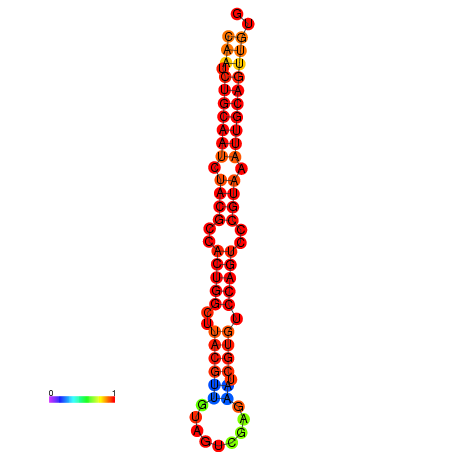 |
| dG=-22.5, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-21.6, p-value=0.009901 |
|---|---|---|
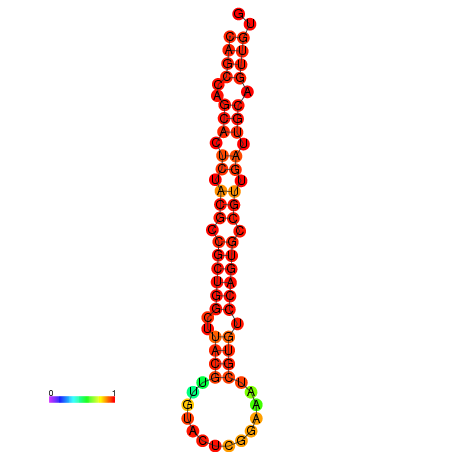 |
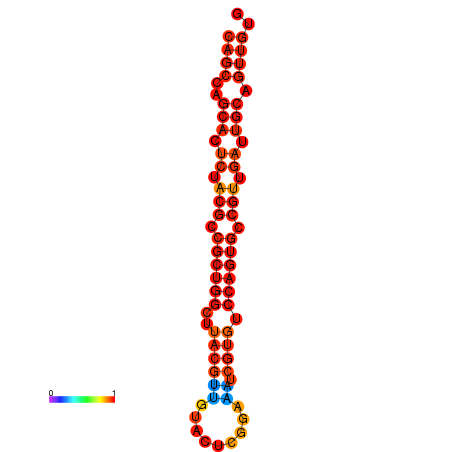 |
 |
| dG=-20.2, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.0, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
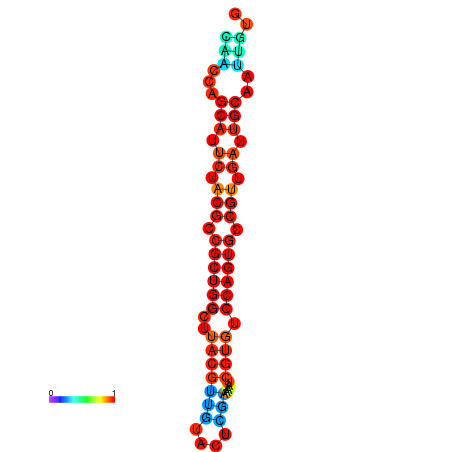 |
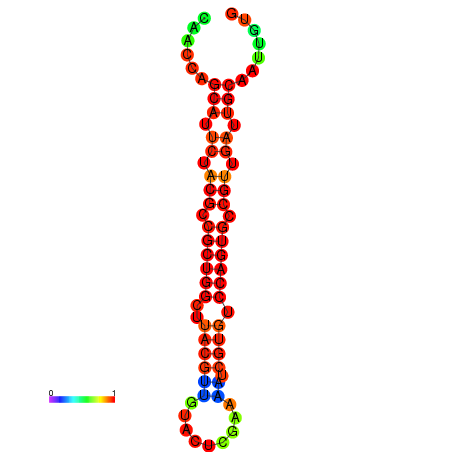 |
| dG=-27.5, p-value=0.009901 | dG=-27.1, p-value=0.009901 |
|---|---|
 |
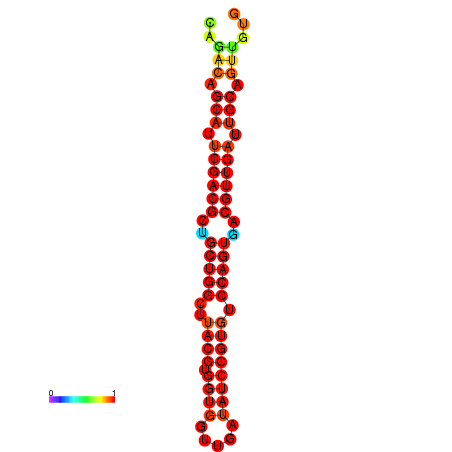 |
No secondary structure predictions returned
No secondary structure predictions returned
| dG=-24.3, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.6, p-value=0.009901 |
|---|---|---|
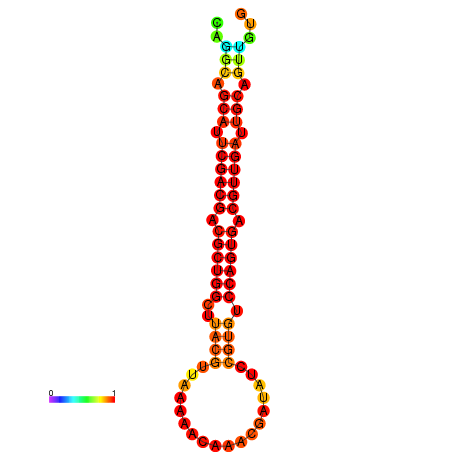 |
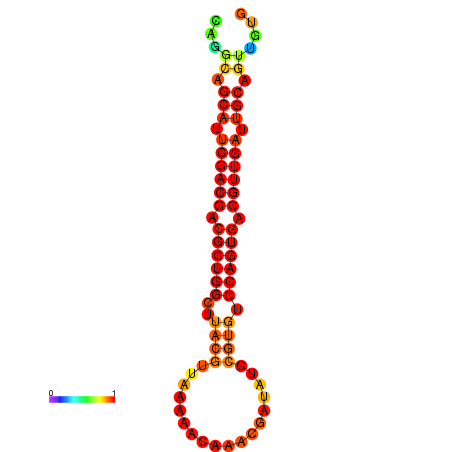 |
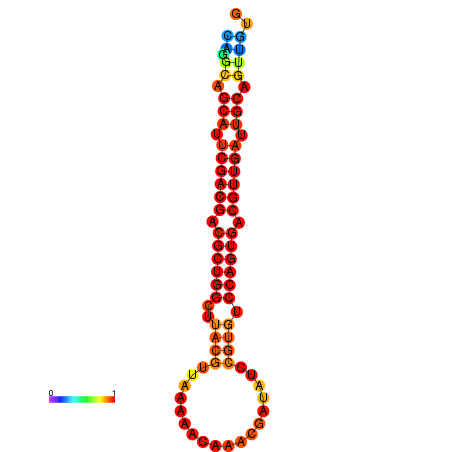 |
| dG=-25.9, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
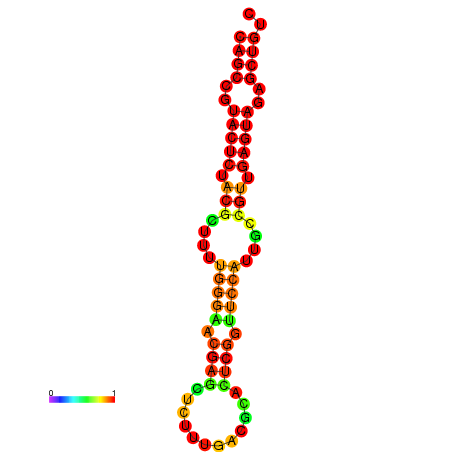 |
 |
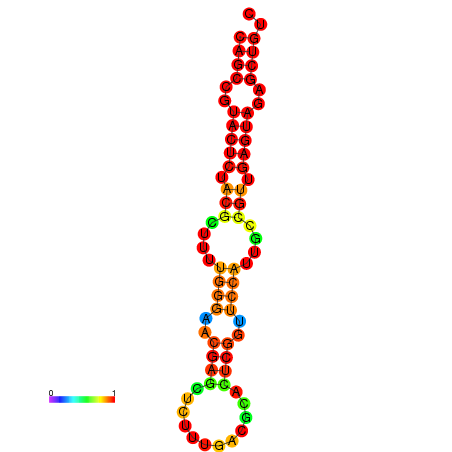 |
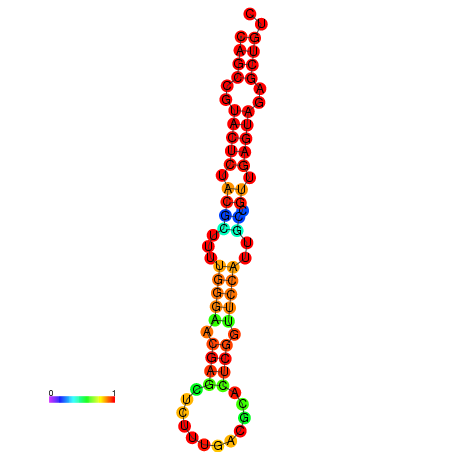 |
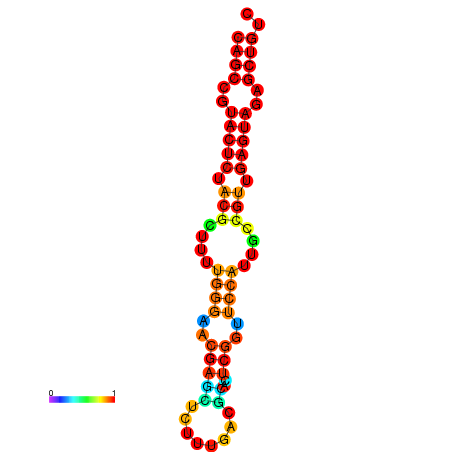 |
| dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
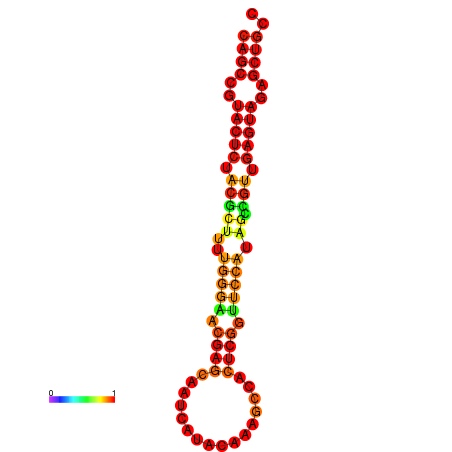 |
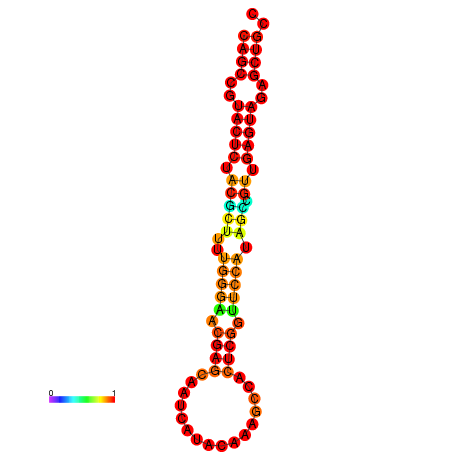 |
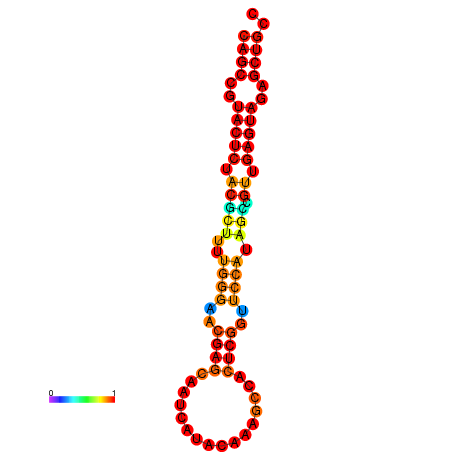 |
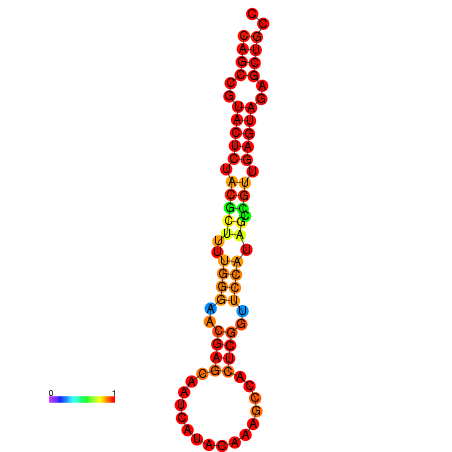 |
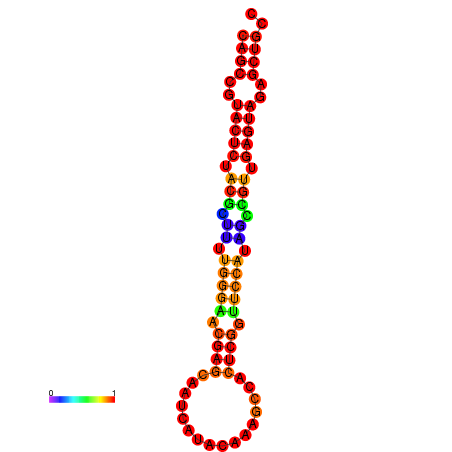 |
| dG=-27.8, p-value=0.009901 | dG=-27.3, p-value=0.009901 |
|---|---|
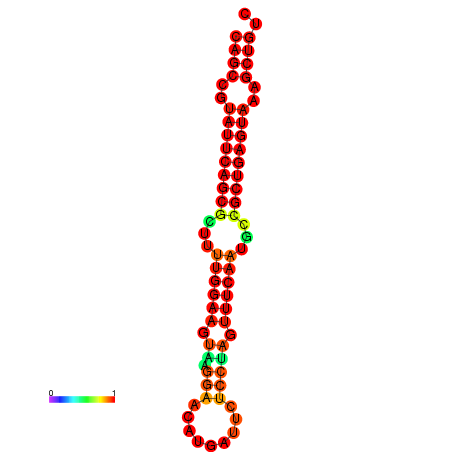 |
 |
Generated: 03/07/2013 at 07:18 PM