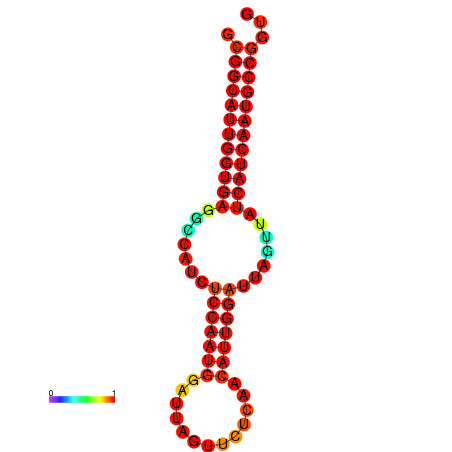
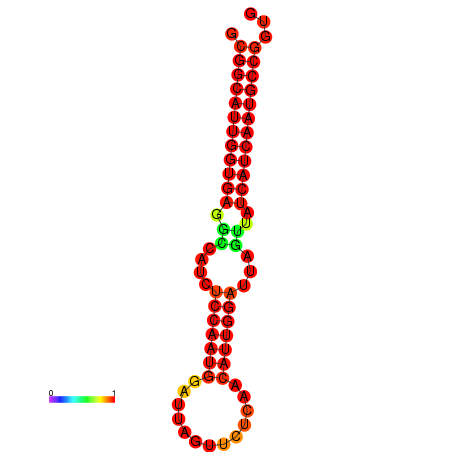
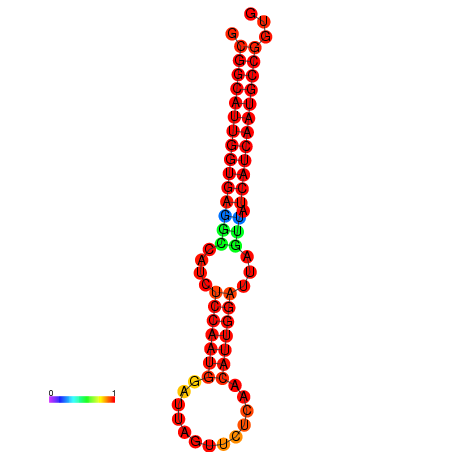
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19452913-19453039 + | AAC------------------G-----GCGGTG-GTGGGCAT--------------------------CG-----CCATGCAGTGCCGCGGCATTGGTGAGGC-CATCTCCAATGG-ATTAGTTCT---CA------ACATTGGATTAGTTATCATCAATGCCGGTGCACTGCACC--TAACAACATC---------------GCGCCGG |
| droSim1 | chrX:15018511-15018617 + | -----------------------------------------------------------------------------ATGCAGTGTCGTGGCATTGGTGAGGA-CATCTCCAATGG-ATTGGTTCT---CA------TCATTGGATTAGTTATCATCAATGCCGGTGCACTGCACTGCTGTCAACTTC---------------GCGCAAT |
| droSec1 | super_8:1735239-1735349 + | TCC------------------G------------------------------------------------------CATGCAGTGCCGTGGCATTGGTGAGGA-CATCTCCAATGG-CTTAGTTCT---CA------TCATTGGATTAGTTATCATCAATGCCGGTGCACTGCAGC--TGTCAACTTC---------------GCGCAAT |
| droYak2 | chrX:11885798-11885920 - | AGA------------------G--------GTGAGTGAGAAT--------------------------CG------CATGCAGTGCCACGACATTGGTGAGGG-CATCTCCAATGG-----GGACCCGGCA------TCATTGGATTAGTTCTCATTAATGCCGGAGCACTGCACC--TACCAACTTC---------------GCACGGT |
| droEre2 | scaffold_4690:9688892-9689017 + | TCC------------------A-----GAGGTGTGCCAGAAT--------------------------CG------CATGCAGTGCCTCGACATTGGTGAGGG-CATCTCCAATGG--GGAGCCAG---CA------TCATTGGATTAGTTCTCATTAATGCCGGAGCATGGCACC--TACGAACTGA---------------GCATTTG |
| droAna3 | scaffold_13417:2467561-2467679 - | GCT------------------A-----CCGATG-------------------------------------CTCCCCGAAGGAGCGCCTCGACATTGGTGAGTT-CATCTCCAATGG--TCAGTAGC---CC------GCATTGGATTAGTTCTCATTGATGGCGAAGCGAGT--CC--TACCCCCACA---------------CCAGCGA |
| dp4 | Unknown | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droPer1 | Unknown | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droWil1 | scaffold_181096:7956743-7956912 + | CTCTCCCCTTCCTTCTTACACA-----GGATTATG-TAGGAT--------------------------TA------T-GGCAACACCTCGATATTGGTGAGTTCCAGCTCCAATGTTATTGGAGCCCAACATTGAACACATTGGATTAGTTCTCATCGATGGCGAAGTGTTT-GTGAGCAATATCGTATATTGTGCGTGCAGTGTATTGT |
| droVir3 | scaffold_13042:906708-906880 - | ATC------------------GTTATCGTTATC-GTTATCGTTATCAGAATTAATTATCGTTGAAGTGCGTTCTTTGGTGCAGCCCCTCGCGATTGATGATAG-CATCTCCAATGA-----GCATA---TC------ACATTGGATTAGTTCTCATCGATGCCGATGCGCTGTACCAGCAACAACAAC---AGGAATAGCAACGGAAACA |
| droMoj3 | scaffold_6328:2900970-2901098 - | TAT------------------C-----GTTATC-GCTAGCGA--------------------------CGCTTC-GGGTGCAGCCCCTCGCGATTGATGAAAG-CATCTCCAATGA-----GTATT---TC------GCATTGGATTAGTTCTCATCGATGTCGATGCGCTGTATCAGCAACAACAAC---------------AACATGA |
| droGri2 | Unknown | ------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19452913-19453039 + | AAC------------------G-----GCGGTG-GTGGGCAT--------------------------CG-----CCATGCAGTGCCGCGGCATTGGTGAGGC-CATCTCCAATGG-ATTAGTTCT---CA------ACATTGGATTAGTTATCATCAATGCCGGTGCACTGCACC--TAACAACATC---------------GCGCCGG |
| droSim1 | chrX:15018511-15018617 + | -----------------------------------------------------------------------------ATGCAGTGTCGTGGCATTGGTGAGGA-CATCTCCAATGG-ATTGGTTCT---CA------TCATTGGATTAGTTATCATCAATGCCGGTGCACTGCACTGCTGTCAACTTC---------------GCGCAAT |
| droSec1 | super_8:1735239-1735349 + | TCC------------------G------------------------------------------------------CATGCAGTGCCGTGGCATTGGTGAGGA-CATCTCCAATGG-CTTAGTTCT---CA------TCATTGGATTAGTTATCATCAATGCCGGTGCACTGCAGC--TGTCAACTTC---------------GCGCAAT |
| droYak2 | chrX:11885798-11885920 - | AGA------------------G--------GTGAGTGAGAAT--------------------------CG------CATGCAGTGCCACGACATTGGTGAGGG-CATCTCCAATGG-----GGACCCGGCA------TCATTGGATTAGTTCTCATTAATGCCGGAGCACTGCACC--TACCAACTTC---------------GCACGGT |
| droEre2 | scaffold_4690:9688892-9689017 + | TCC------------------A-----GAGGTGTGCCAGAAT--------------------------CG------CATGCAGTGCCTCGACATTGGTGAGGG-CATCTCCAATGG--GGAGCCAG---CA------TCATTGGATTAGTTCTCATTAATGCCGGAGCATGGCACC--TACGAACTGA---------------GCATTTG |
| droAna3 | scaffold_13417:2467561-2467679 - | GCT------------------A-----CCGATG-------------------------------------CTCCCCGAAGGAGCGCCTCGACATTGGTGAGTT-CATCTCCAATGG--TCAGTAGC---CC------GCATTGGATTAGTTCTCATTGATGGCGAAGCGAGT--CC--TACCCCCACA---------------CCAGCGA |
| droWil1 | scaffold_181096:7956743-7956912 + | CTCTCCCCTTCCTTCTTACACA-----GGATTATG-TAGGAT--------------------------TA------T-GGCAACACCTCGATATTGGTGAGTTCCAGCTCCAATGTTATTGGAGCCCAACATTGAACACATTGGATTAGTTCTCATCGATGGCGAAGTGTTT-GTGAGCAATATCGTATATTGTGCGTGCAGTGTATTGT |
| droVir3 | scaffold_13042:906708-906880 - | ATC------------------GTTATCGTTATC-GTTATCGTTATCAGAATTAATTATCGTTGAAGTGCGTTCTTTGGTGCAGCCCCTCGCGATTGATGATAG-CATCTCCAATGA-----GCATA---TC------ACATTGGATTAGTTCTCATCGATGCCGATGCGCTGTACCAGCAACAACAAC---AGGAATAGCAACGGAAACA |
| droMoj3 | scaffold_6328:2900970-2901098 - | TAT------------------C-----GTTATC-GCTAGCGA--------------------------CGCTTC-GGGTGCAGCCCCTCGCGATTGATGAAAG-CATCTCCAATGA-----GTATT---TC------GCATTGGATTAGTTCTCATCGATGTCGATGCGCTGTATCAGCAACAACAAC---------------AACATGA |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
| dG=-28.6, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.3, p-value=0.009901 |
|---|---|---|
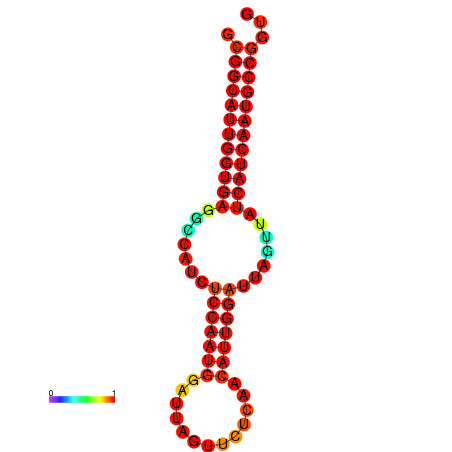 |
 |
 |
| dG=-28.4, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.7, p-value=0.009901 |
|---|---|---|---|---|
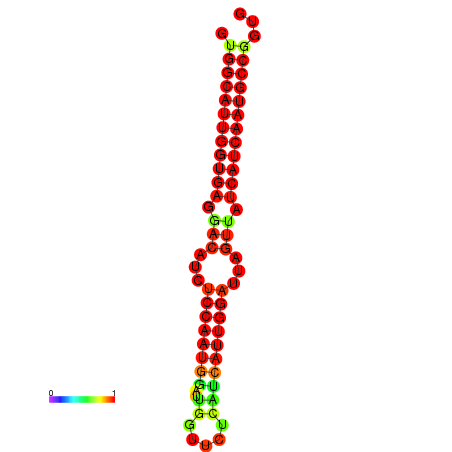 |
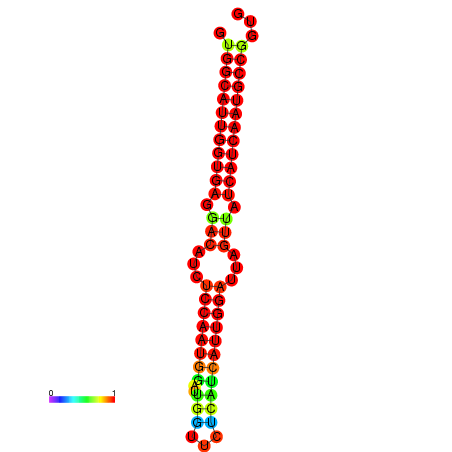 |
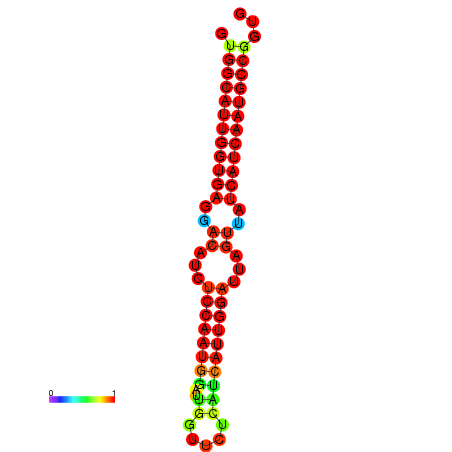 |
 |
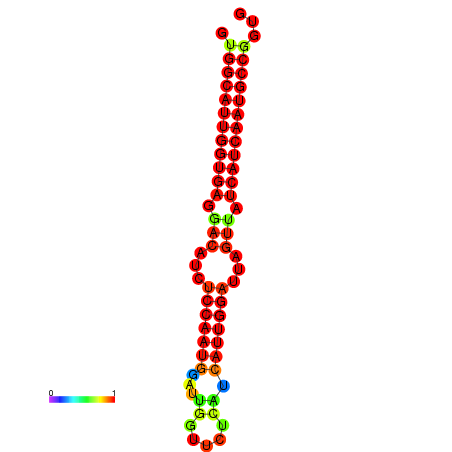 |
| dG=-27.8, p-value=0.009901 | dG=-27.5, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-27.0, p-value=0.009901 |
|---|---|---|---|---|
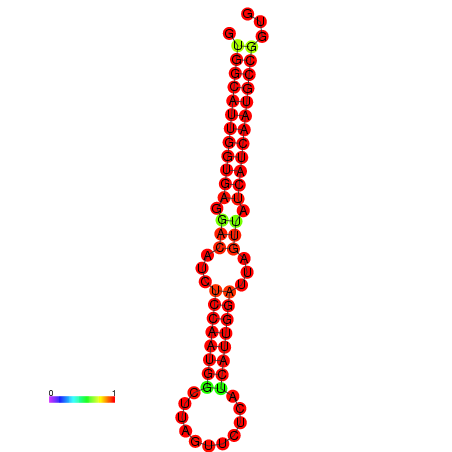 |
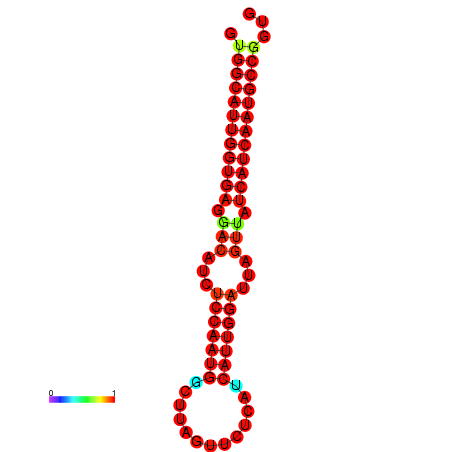 |
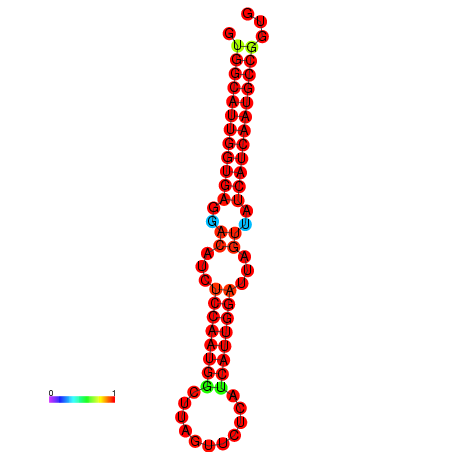 |
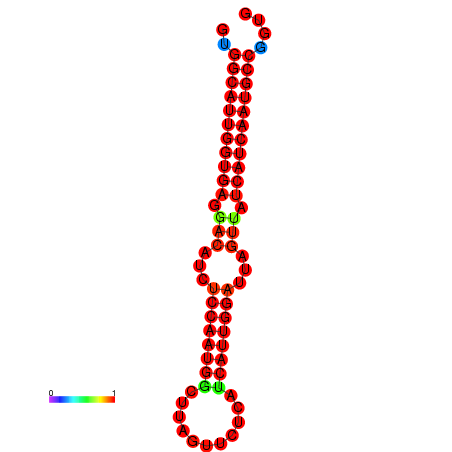 |
 |
| dG=-26.6, p-value=0.009901 |
|---|
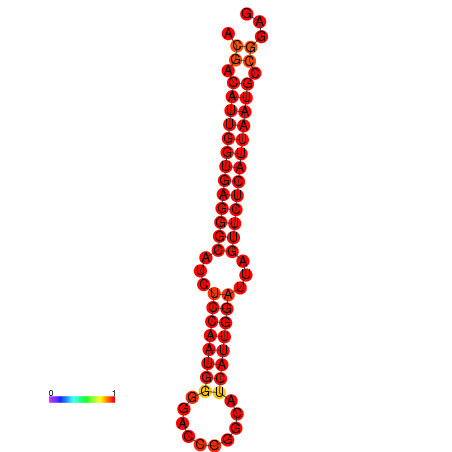 |
| dG=-26.9, p-value=0.009901 | dG=-26.2, p-value=0.009901 |
|---|---|
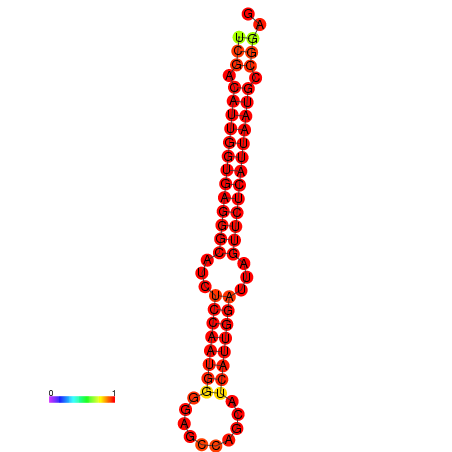 |
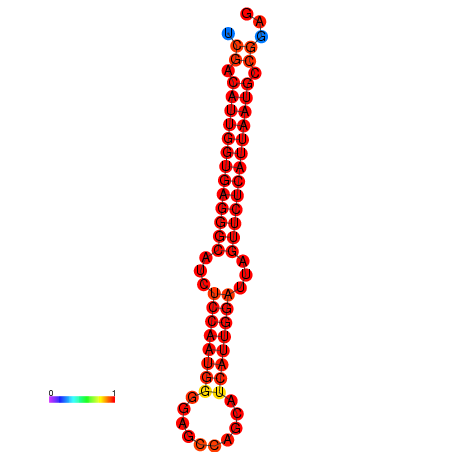 |
| dG=-21.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 |
|---|---|
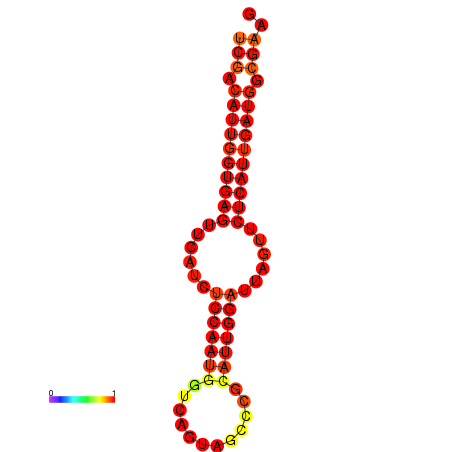 |
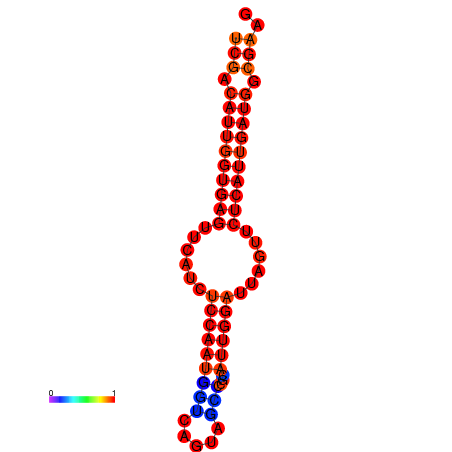 |
No secondary structure predictions returned
No secondary structure predictions returned
| dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|---|---|---|
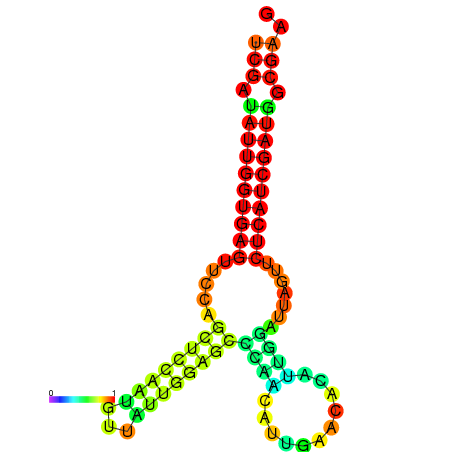 |
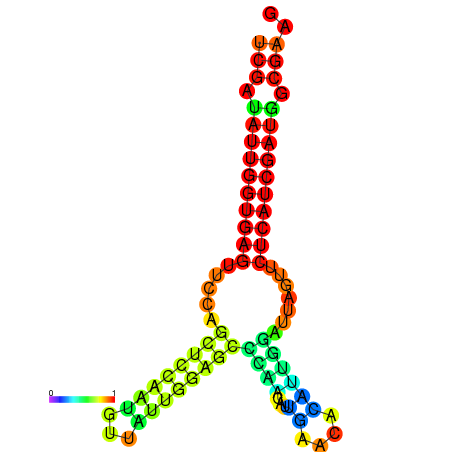 |
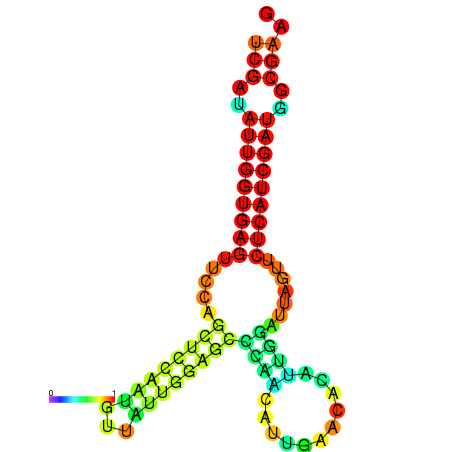 |
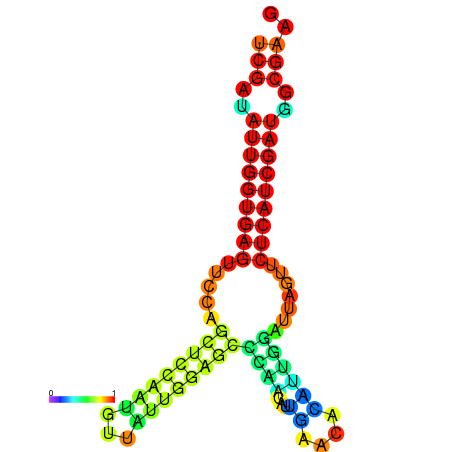 |
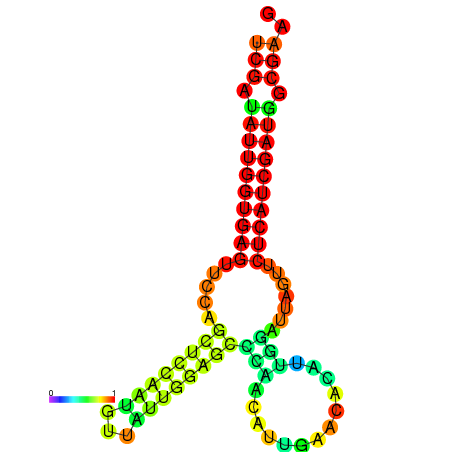 |
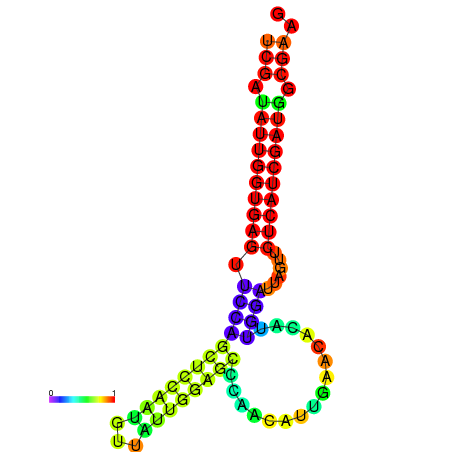 |
| dG=-21.0, p-value=0.009901 | dG=-20.0, p-value=0.009901 |
|---|---|
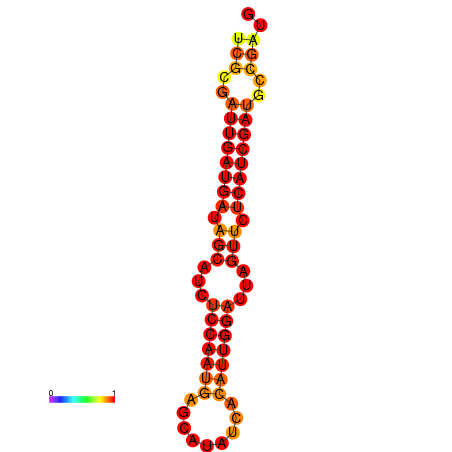 |
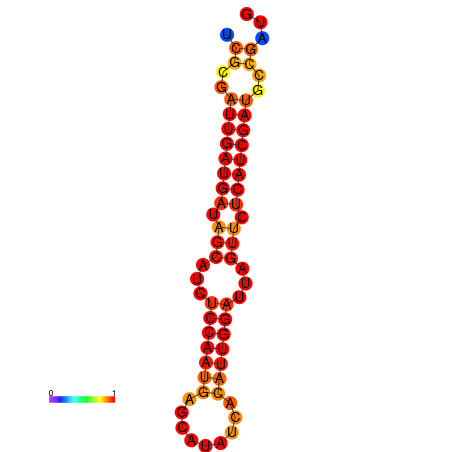 |
| dG=-21.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 |
|---|---|
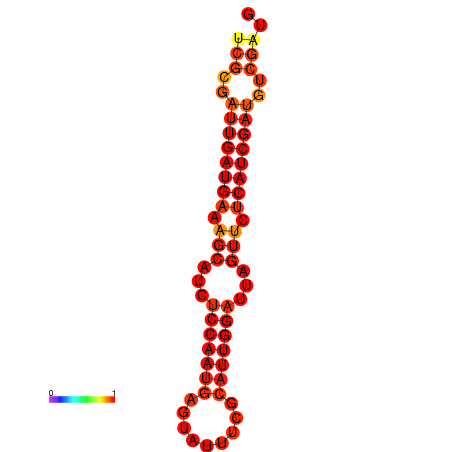 |
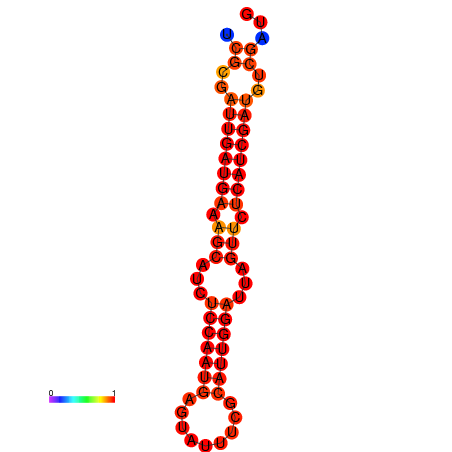 |
No secondary structure predictions returned
Generated: 03/07/2013 at 07:17 PM