
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19452771-19452891 + | ATCCAA--------------TTGGCAATATTGAATTTTTGATTTTAAACACTTCCTACATCCTGTATGTGT-TTTG----------------------------CATCCGGTACAGATGTGGGAGTCGTTTGCACTCAGAGATTTCACAGAATTCGAGGATC-GA |
| droSim1 | chrX:15018379-15018489 + | -------------------------AATATTGAGTTTTTGATTTTAAACACTGCCTACATCCTGTATGTGT-TTTG----------------------------CATCCGATACAGTTGTAGGAGTCGTTTGCAGTCAGAGATTTCACAGACTGCGAGGATC-GA |
| droSec1 | super_8:1735110-1735229 + | A-AAA--------------TTAATTAATATTGAGTTTTTGATTTTAAACACTCCCTACATCCTGTATGTGT-TTTG----------------------------CATCCGATACAGTTGTAGGAGTCGTTTGCAGTCAGAGATTTCACAGACTGCGAGGATC-GC |
| droYak2 | chrX:11885955-11886071 - | ATCCA------------------GCAATATTGAATTCTTGACTTTAAACACTGCCTACATACTGTATGTGT-TTTG----------------------------CACCCGATACAGATGTGGGGGTCGTTTGCAATCAGAGATTCCGCAGGCTACGAGGATC-GG |
| droEre2 | scaffold_4690:9688756-9688876 + | -TCCA--------------ACCATCAACATTGAATTCTTGACTTTAAACACTTCCTACATCCTGTATGTGT-TTTG----------------------------CGCCCTATACAGATGTGGGGGTCGTTTGCACTCAGGGATTCCACAGGCTGCAAGGATC-GG |
| droAna3 | scaffold_13417:2467701-2467813 - | GTCCAAG------------CCAGTCGACAATGAATATTTGATTTTAAACACTACCTACATCCAGTATGTTACTTGA----------------------------CTCCCAATACGGATGTGGAGGGCGTTTACACTCAGAGATTCTGGAG-----------C-CA |
| dp4 | Unknown | --------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 | Unknown | --------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 | scaffold_181096:7956608-7956722 + | GTTTTGTAGATGTGTTGGAATC-ACGACATTCAACTGTTATGTTTACATATGTCTAACATTCTGT-----------------TATAGTATAACAGTGTGTCTTGCG----------------------TTCCTACTCAACAC-TCAACAT-TCGC--------AA |
| droVir3 | scaffold_13042:906902-907003 - | ----------------------------------TCTTTGATTTTAGATACTCCCTATATCCAGTATGTGT-GCCACTT-ATTGAAATACA-----------------------AGATGTATGGGGTGTCTGCAATCACAAACCACGTAG-CCGCAA---TCAGC |
| droMoj3 | scaffold_6328:2901122-2901224 - | ----------------------------------TCTTTGATTGTAGATACTCCCTATATCCCGTATGTGC-GATACTTTGATAAAATACA-----------------------AGATGTATGGGGTGTCTGCAATCACAAACCACATAG-CCGCAA---TCAAA |
| droGri2 | Unknown | --------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19452771-19452891 + | ATCCAA--------------TTGGCAATATTGAATTTTTGATTTTAAACACTTCCTACATCCTGTATGTGT-TTTG----------------------------CATCCGGTACAGATGTGGGAGTCGTTTGCACTCAGAGATTTCACAGAATTCGAGGATC-GA |
| droSim1 | chrX:15018379-15018489 + | -------------------------AATATTGAGTTTTTGATTTTAAACACTGCCTACATCCTGTATGTGT-TTTG----------------------------CATCCGATACAGTTGTAGGAGTCGTTTGCAGTCAGAGATTTCACAGACTGCGAGGATC-GA |
| droSec1 | super_8:1735110-1735229 + | A-AAA--------------TTAATTAATATTGAGTTTTTGATTTTAAACACTCCCTACATCCTGTATGTGT-TTTG----------------------------CATCCGATACAGTTGTAGGAGTCGTTTGCAGTCAGAGATTTCACAGACTGCGAGGATC-GC |
| droYak2 | chrX:11885955-11886071 - | ATCCA------------------GCAATATTGAATTCTTGACTTTAAACACTGCCTACATACTGTATGTGT-TTTG----------------------------CACCCGATACAGATGTGGGGGTCGTTTGCAATCAGAGATTCCGCAGGCTACGAGGATC-GG |
| droEre2 | scaffold_4690:9688756-9688876 + | -TCCA--------------ACCATCAACATTGAATTCTTGACTTTAAACACTTCCTACATCCTGTATGTGT-TTTG----------------------------CGCCCTATACAGATGTGGGGGTCGTTTGCACTCAGGGATTCCACAGGCTGCAAGGATC-GG |
| droAna3 | scaffold_13417:2467701-2467813 - | GTCCAAG------------CCAGTCGACAATGAATATTTGATTTTAAACACTACCTACATCCAGTATGTTACTTGA----------------------------CTCCCAATACGGATGTGGAGGGCGTTTACACTCAGAGATTCTGGAG-----------C-CA |
| droWil1 | scaffold_181096:7956608-7956722 + | GTTTTGTAGATGTGTTGGAATC-ACGACATTCAACTGTTATGTTTACATATGTCTAACATTCTGT-----------------TATAGTATAACAGTGTGTCTTGCG----------------------TTCCTACTCAACAC-TCAACAT-TCGC--------AA |
| droVir3 | scaffold_13042:906902-907003 - | ----------------------------------TCTTTGATTTTAGATACTCCCTATATCCAGTATGTGT-GCCACTT-ATTGAAATACA-----------------------AGATGTATGGGGTGTCTGCAATCACAAACCACGTAG-CCGCAA---TCAGC |
| droMoj3 | scaffold_6328:2901122-2901224 - | ----------------------------------TCTTTGATTGTAGATACTCCCTATATCCCGTATGTGC-GATACTTTGATAAAATACA-----------------------AGATGTATGGGGTGTCTGCAATCACAAACCACATAG-CCGCAA---TCAAA |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
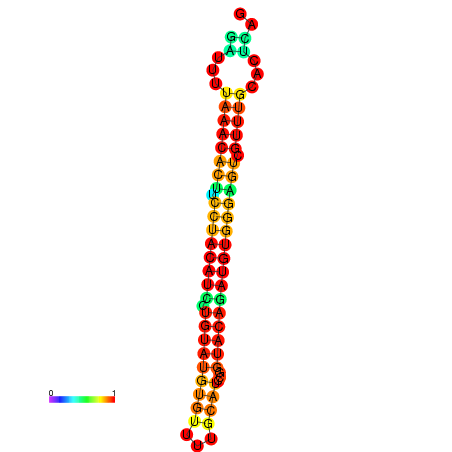
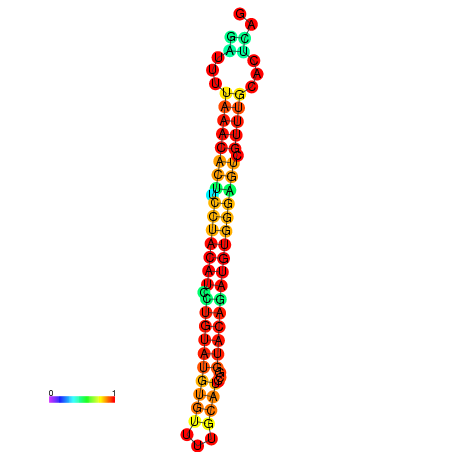
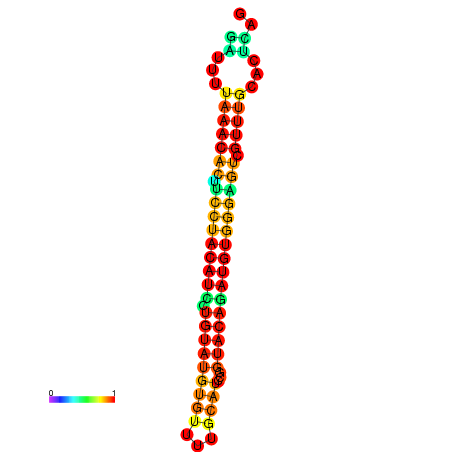
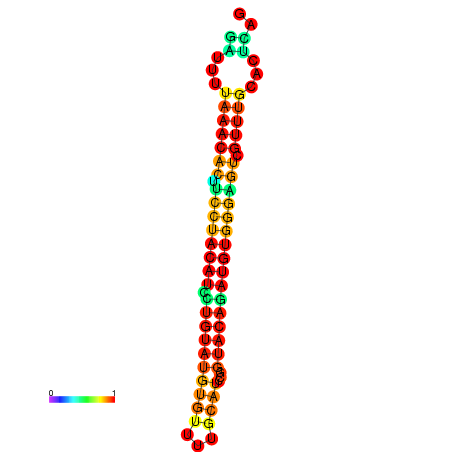
| dG=-21.7, p-value=0.009901 | dG=-21.7, p-value=0.009901 | dG=-21.7, p-value=0.009901 | dG=-21.7, p-value=0.009901 | dG=-21.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-23.3, p-value=0.009901 |
|---|
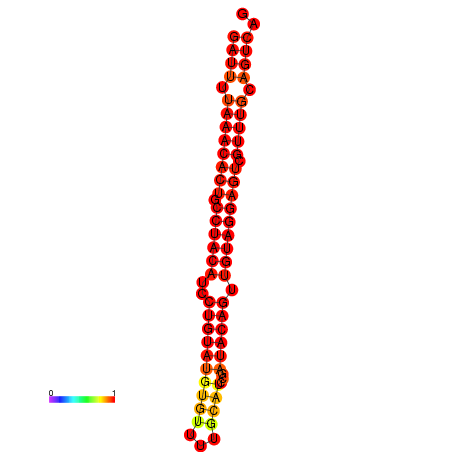 |
| dG=-23.3, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|
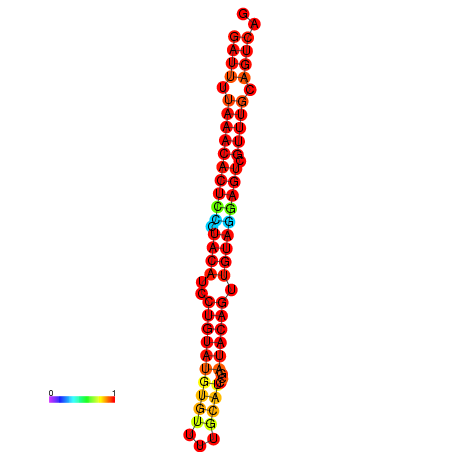 |
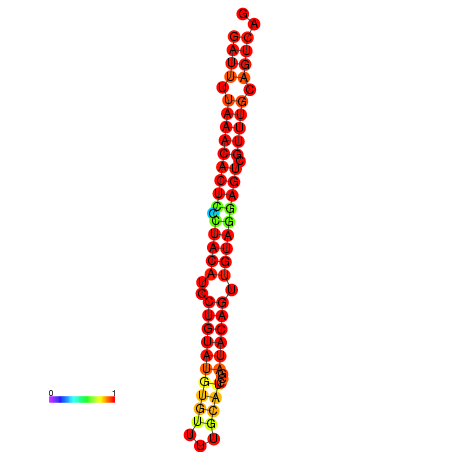 |
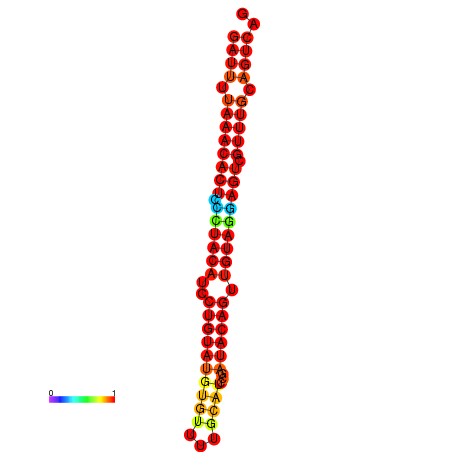 |
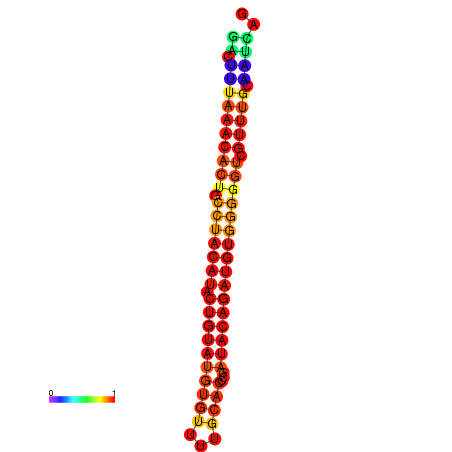
| dG=-21.8, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-20.9, p-value=0.009901 | dG=-20.9, p-value=0.009901 | dG=-20.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
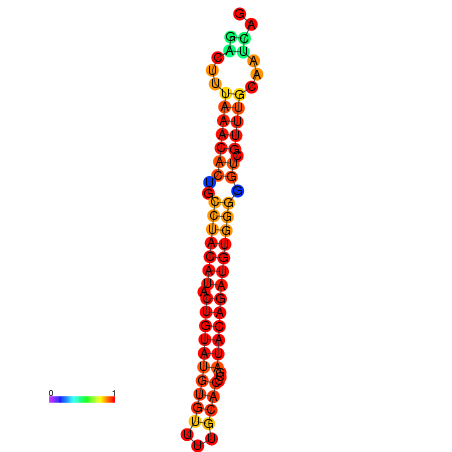 |
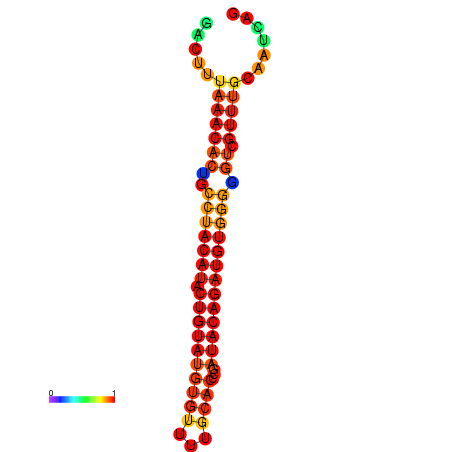 |
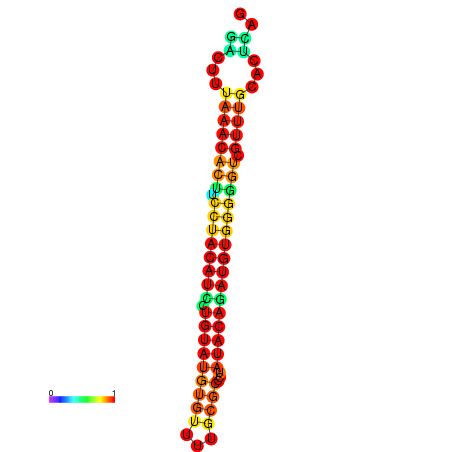
| dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
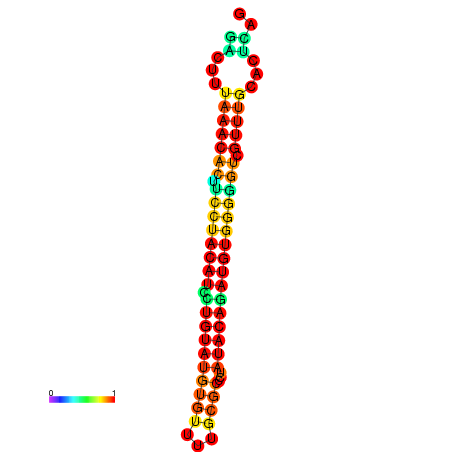 |
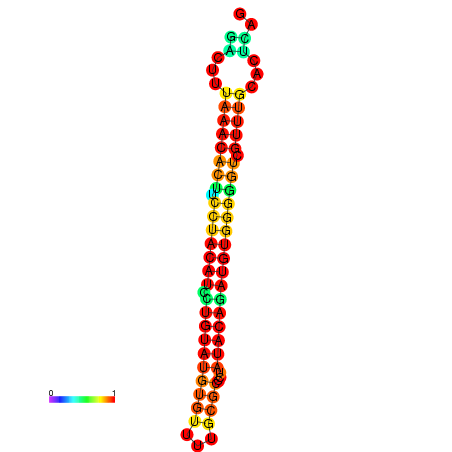
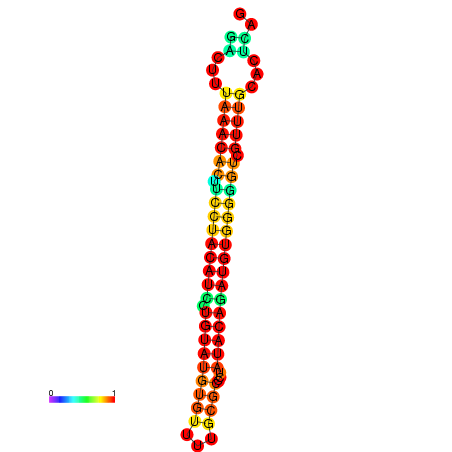
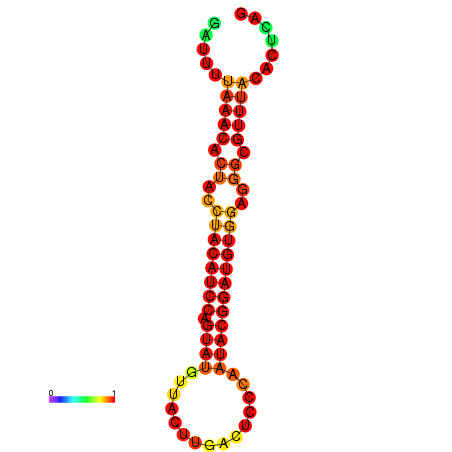
| dG=-17.8, p-value=0.009901 | dG=-17.8, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.9, p-value=0.009901 |
|---|---|---|---|
 |
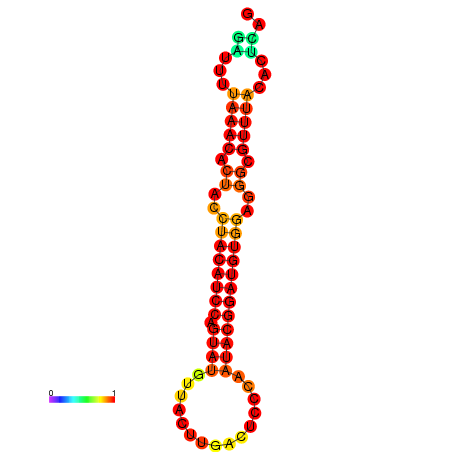 |
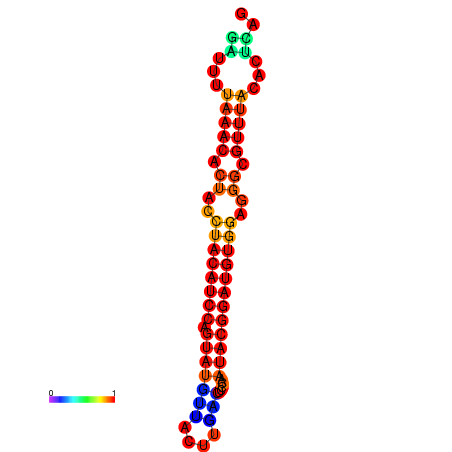 |
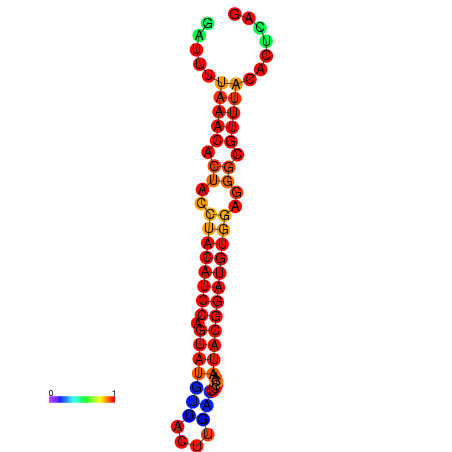 |
No secondary structure predictions returned
No secondary structure predictions returned
| dG=-11.0, p-value=0.009901 | dG=-10.9, p-value=0.009901 | dG=-10.9, p-value=0.009901 | dG=-10.2, p-value=0.009901 | dG=-10.1, p-value=0.009901 |
|---|---|---|---|---|
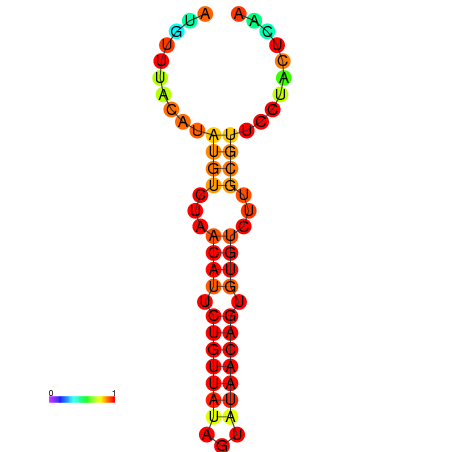 |
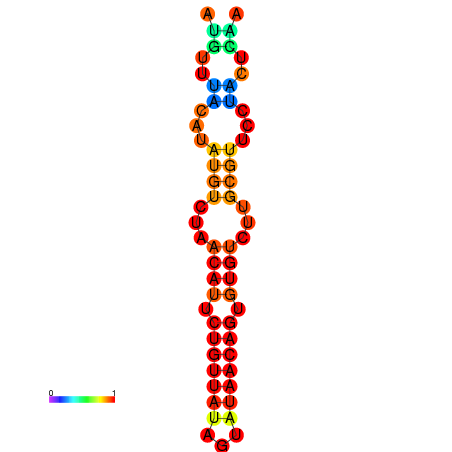 |
 |
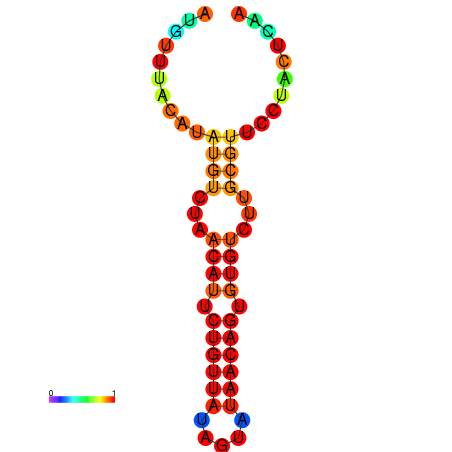 |
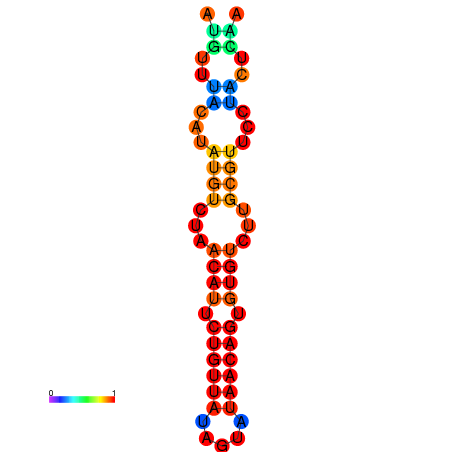 |
| dG=-25.4, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|
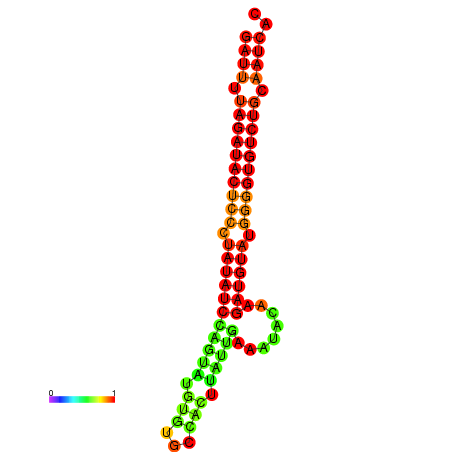 |
 |
| dG=-32.1, p-value=0.009901 | dG=-31.6, p-value=0.009901 |
|---|---|
 |
 |
No secondary structure predictions returned
Generated: 03/07/2013 at 07:17 PM