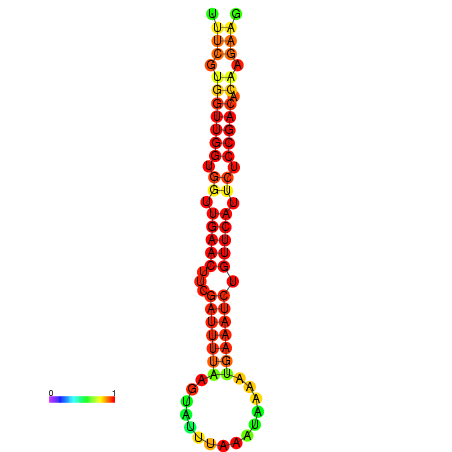
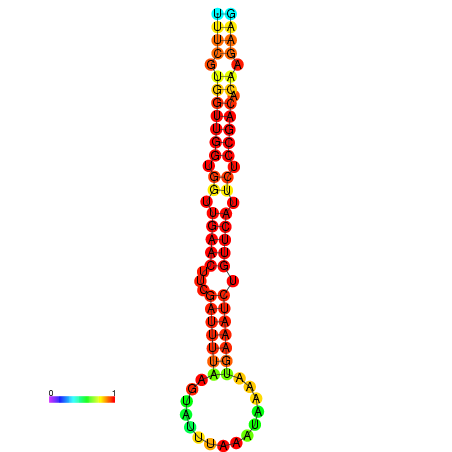
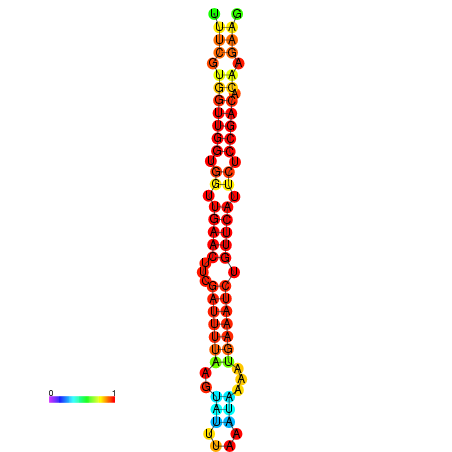
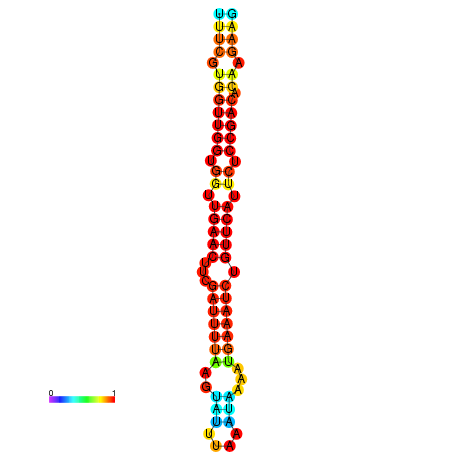
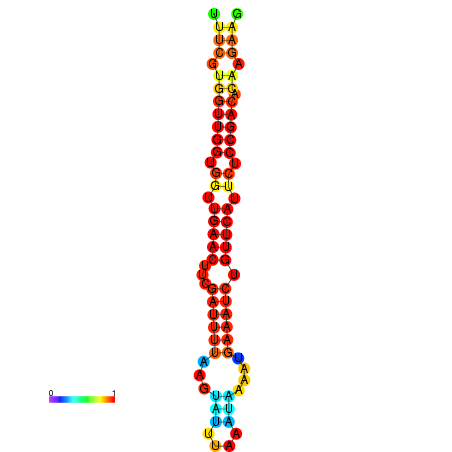
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19445546-19445676 + | AATATACTTC----ACTTTA-CCGTCGTCGACTTTTCGTGGTTGGTGGTTGAACTTCGATTTTAAG-TA--TTT--------AAATAAAATGAAATCTGTTCATTCTCCGACACAAGAAGTTCACGCAAAGGA------------------------GCTGCAGCTGCGAA |
| droSim1 | chrX:15011303-15011407 + | CG-------------------------TATACTTTTCCGGGTTGGTGGTTGAACTTCGATTTTAAG-CA--TTT--------AAATAAGA-TAAATCTGTTCATTCTTCGACCCAAAAAGTTCACGTGAAGAA------------------------GCAGCG------AA |
| droSec1 | super_8:1727974-1728085 + | AC------------------TTCAACGTATACTTTTCGGGGTTGGTGGTTGAACTTCGATTTTAGG-TA--TTT--------AAATAAGA-TAAATCTGTTCATTCTTCGACCCAAAAAGTTCACGTGAAGAA------------------------GCAGCG------AG |
| droYak2 | Unknown | GTTATAA--G----AT----TTTAACGTCGACTTTTCGTGGTTGGTGGCTGAACTTAGATTTTGAG-TA--TAC--------AAATAGAA-CAAATCTGTTCATTCCTCGACACAACAAGTTGACGTAAAGGA------------------------GCAGCA------AA |
| droEre2 | scaffold_4690:9682299-9682417 + | ACTATAC--C----AT----TTTAACGTCGACTTTTCGTGGTTGGTGGCTGAACTTAGATTTTGAG-TA--TTC--------AAATAAAA-TAAATCTGTTCATTCCTCGACACAAGAAGTTGACGTGCAGGA------------------------GCGGCA------AA |
| droAna3 | scaffold_13417:2478622-2478750 - | -----------------------GACACCGACTCCTCGTGTTTGGCGATTGAACCTGGATTTTTAA-TT--TTC--------GAGTACGA-GAAATCTGTTCATTCCTCGACACTAGTAGTTGGCGCGACA-C------TCAGCGCACCAAACATGAACCGCGGCTCAAAT |
| dp4 | Unknown | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 | Unknown | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 | scaffold_181096:7946449-7946592 + | AAAATATATCGGGAATATTG-TCGACATGGCCTTTTCGTGTTTGGTGATTGAACTTGGATTTTGTTCTATATACATATATAAAAAT-ATG-TAAATCTGTTCATTCCTCGACACTAGAAGTTCATGTCAATAACTTCATTT----------------CCTGCA-------- |
| droVir3 | scaffold_13042:918337-918453 - | AGAACAGTTA----AAATCATCGGACATTCACTTTTCGTGTTCGATGTTTGAACTTGGATTTTTTT-TA--TAT--------A-----CA-CAAATCTGTTCATTCCTCGACACTAGAAGTTTGCGTCAATAA------------------------GCTGC--------- |
| droMoj3 | scaffold_6328:2909116-2909204 - | ----------------------CGGCTTTCACTTTTCGTGTTCGATGTTTGAACTTGGATTTTTAA-AA--TAT--------GAA---AA-GAAATCTGTTCATTCCTCGACACTAAGAGTTTAGG--------------------------------------------- |
| droGri2 | scaffold_15081:2051526-2051596 - | -----------------------GACATTTACTTTTTGTGTTGGGTGATTC---------------------------------------------TGATTCACTTTTCGACACATCAAGTTCACCTCAATAA------------------------ACTGCA-------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19445546-19445676 + | AATATACTTC----ACTTTA-CCGTCGTCGACTTTTCGTGGTTGGTGGTTGAACTTCGATTTTAAG-TA--TTT--------AAATAAAATGAAATCTGTTCATTCTCCGACACAAGAAGTTCACGCAAAGGA------------------------GCTGCAGCTGCGAA |
| droSim1 | chrX:15011303-15011407 + | CGTAT-------------------------ACTTTTCCGGGTTGGTGGTTGAACTTCGATTTTAAG-CA--TTT--------AAATAAGA-TAAATCTGTTCATTCTTCGACCCAAAAAGTTCACGTGAAGAA------------------------GCAGCG------AA |
| droSec1 | super_8:1727974-1728085 + | AC------------------TTCAACGTATACTTTTCGGGGTTGGTGGTTGAACTTCGATTTTAGG-TA--TTT--------AAATAAGA-TAAATCTGTTCATTCTTCGACCCAAAAAGTTCACGTGAAGAA------------------------GCAGCG------AG |
| droYak2 | Unknown | GTTATAA--G----AT----TTTAACGTCGACTTTTCGTGGTTGGTGGCTGAACTTAGATTTTGAG-TA--TAC--------AAATAGAA-CAAATCTGTTCATTCCTCGACACAACAAGTTGACGTAAAGGA------------------------GCAGCA------AA |
| droEre2 | scaffold_4690:9682299-9682417 + | ACTATAC--C----AT----TTTAACGTCGACTTTTCGTGGTTGGTGGCTGAACTTAGATTTTGAG-TA--TTC--------AAATAAAA-TAAATCTGTTCATTCCTCGACACAAGAAGTTGACGTGCAGGA------------------------GCGGCA------AA |
| droAna3 | scaffold_13417:2478622-2478750 - | G-----------------------ACACCGACTCCTCGTGTTTGGCGATTGAACCTGGATTTTTAA-TT--TTC--------GAGTACGA-GAAATCTGTTCATTCCTCGACACTAGTAGTTGGCGCGACA-C------TCAGCGCACCAAACATGAACCGCGGCTCAAAT |
| droWil1 | scaffold_181096:7946449-7946592 + | AAAATATATCGGGAATATTG-TCGACATGGCCTTTTCGTGTTTGGTGATTGAACTTGGATTTTGTTCTATATACATATATAAAAAT-ATG-TAAATCTGTTCATTCCTCGACACTAGAAGTTCATGTCAATAACTTCATTT----------------CCTGC--------A |
| droVir3 | scaffold_13042:918337-918453 - | AGAACAGTTA----AAATCATCGGACATTCACTTTTCGTGTTCGATGTTTGAACTTGGATTTTTTT-TA--TAT--------A-----CA-CAAATCTGTTCATTCCTCGACACTAGAAGTTTGCGTCAATAA------------------------GCTGC--------- |
| droMoj3 | scaffold_6328:2909116-2909204 - | ----------------------CGGCTTTCACTTTTCGTGTTCGATGTTTGAACTTGGATTTTTAA-AA--TAT--------GAA---AA-GAAATCTGTTCATTCCTCGACACTAAGAGTTTAGG--------------------------------------------- |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
| dG=-18.8, p-value=0.009901 | dG=-18.7, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-18.3, p-value=0.009901 | dG=-18.1, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-17.7, p-value=0.009901 | dG=-17.1, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.7, p-value=0.009901 | dG=-16.7, p-value=0.009901 |
|---|---|---|---|---|
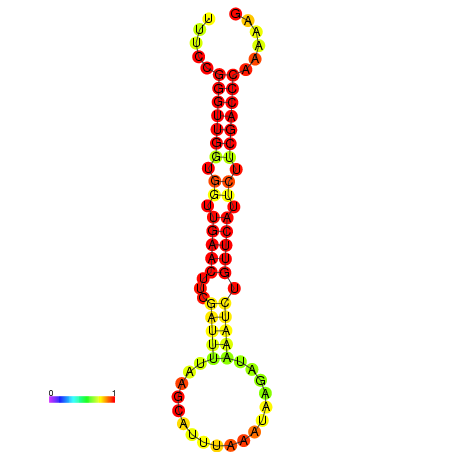 |
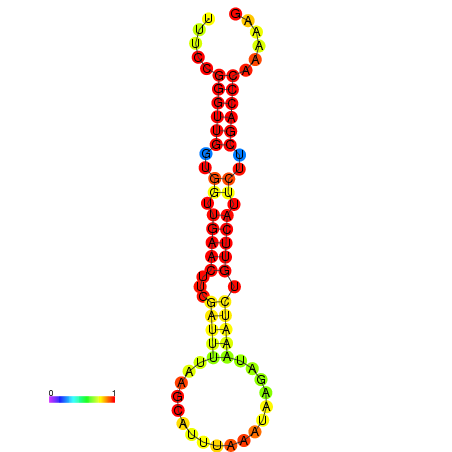 |
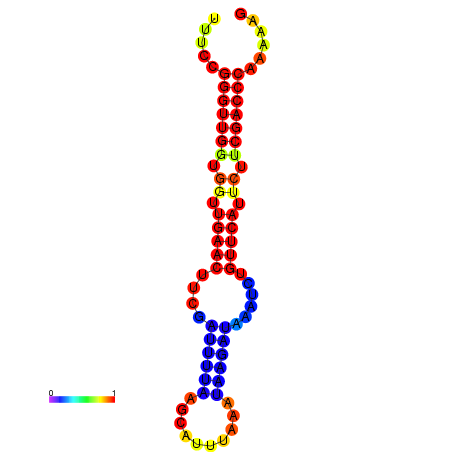 |
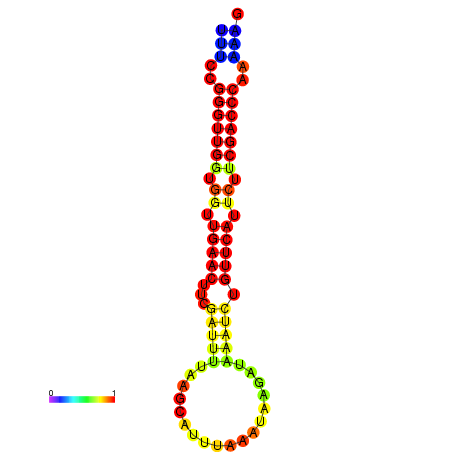 |
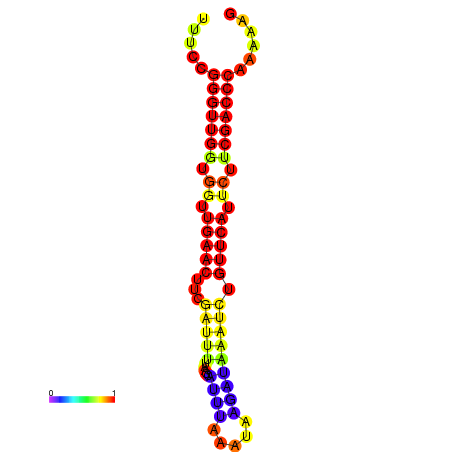 |
| dG=-19.1, p-value=0.009901 | dG=-18.9, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.3, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|---|---|
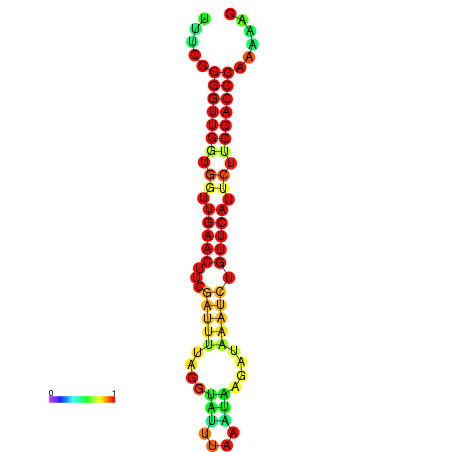 |
 |
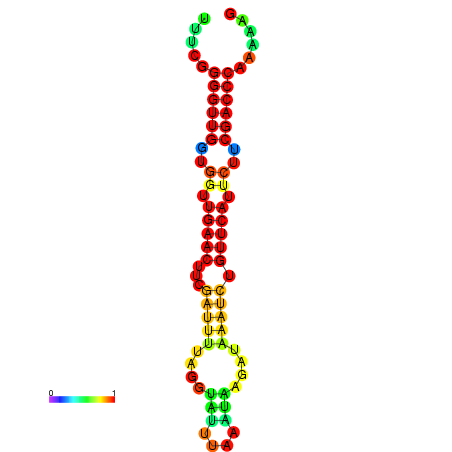 |
 |
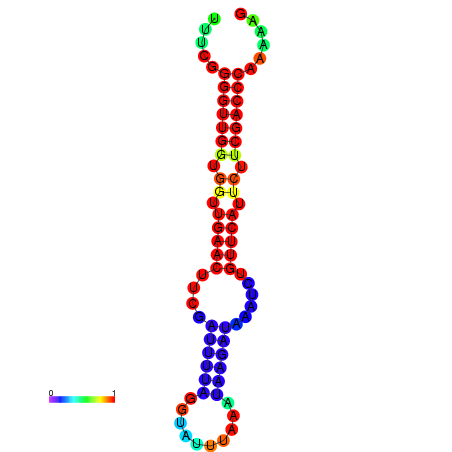 |
| dG=-13.5, p-value=0.009901 | dG=-13.3, p-value=0.009901 | dG=-12.9, p-value=0.009901 | dG=-12.9, p-value=0.009901 | dG=-12.9, p-value=0.009901 |
|---|---|---|---|---|
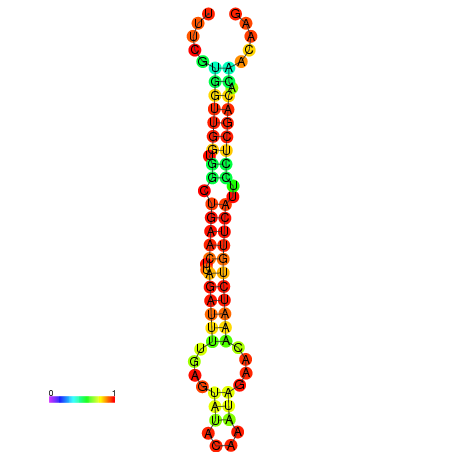 |
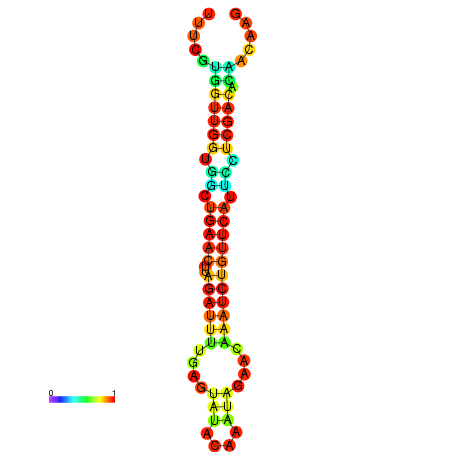 |
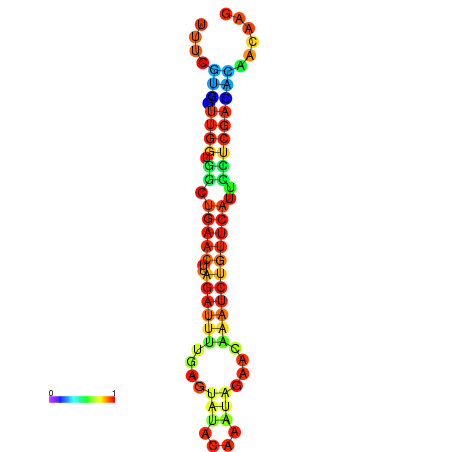 |
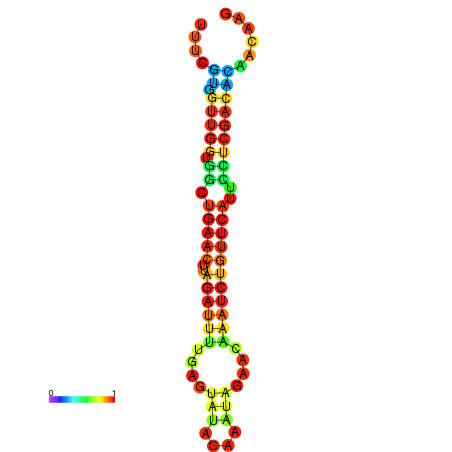 |
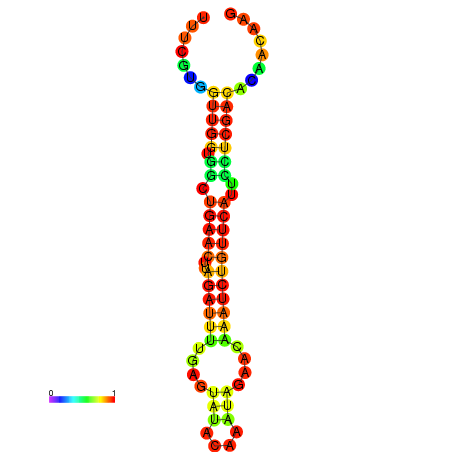 |
| dG=-15.3, p-value=0.009901 | dG=-15.2, p-value=0.009901 | dG=-15.1, p-value=0.009901 | dG=-15.0, p-value=0.009901 | dG=-14.4, p-value=0.009901 |
|---|---|---|---|---|
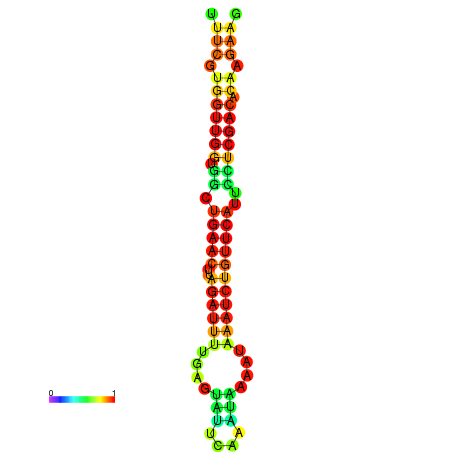 |
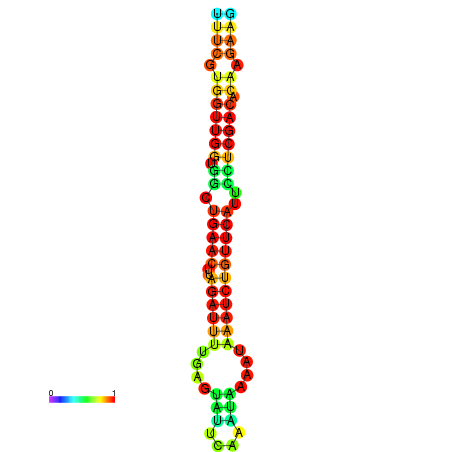 |
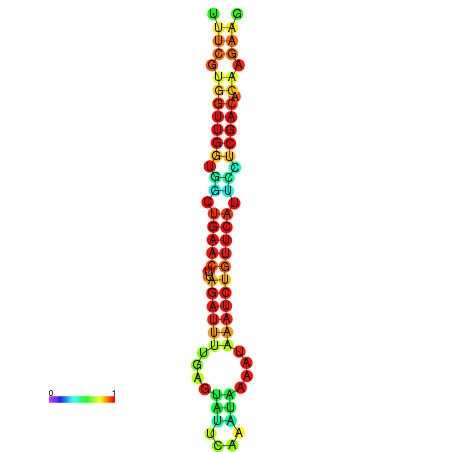 |
 |
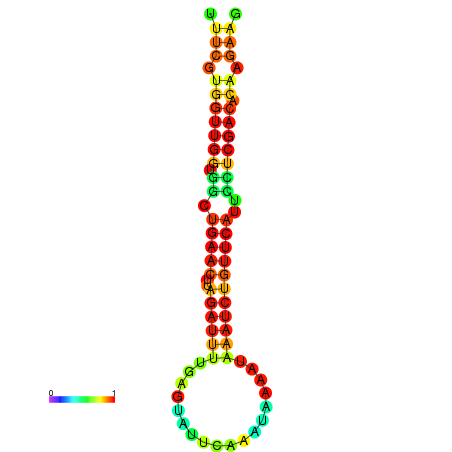 |
| dG=-18.3, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-18.0, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.6, p-value=0.009901 |
|---|---|---|---|---|
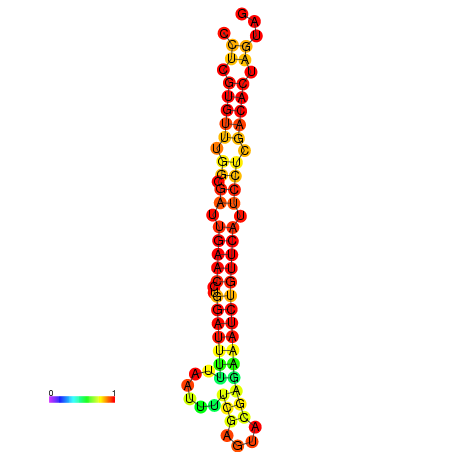 |
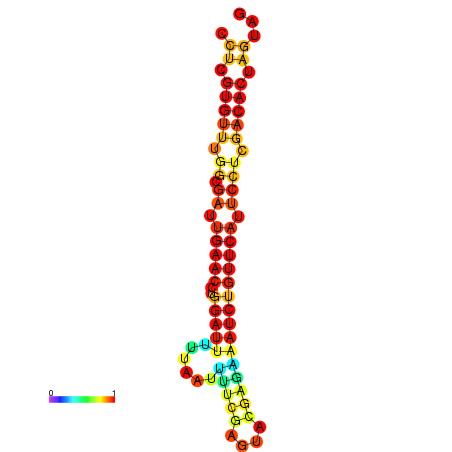 |
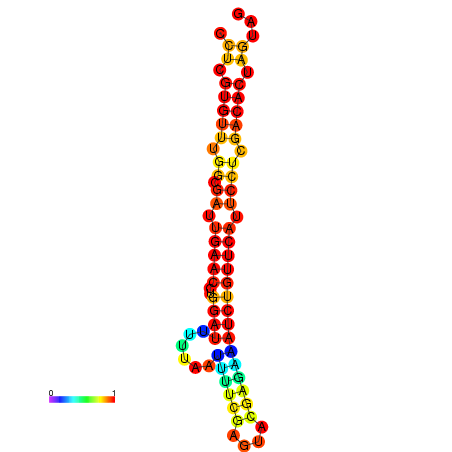 |
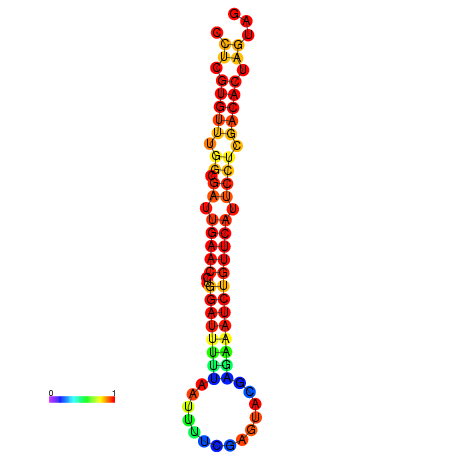 |
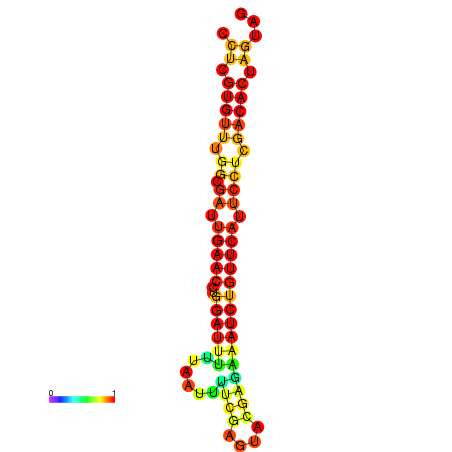 |
No secondary structure predictions returned
No secondary structure predictions returned
| dG=-18.7, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.4, p-value=0.009901 | dG=-18.3, p-value=0.009901 | dG=-18.3, p-value=0.009901 |
|---|---|---|---|---|
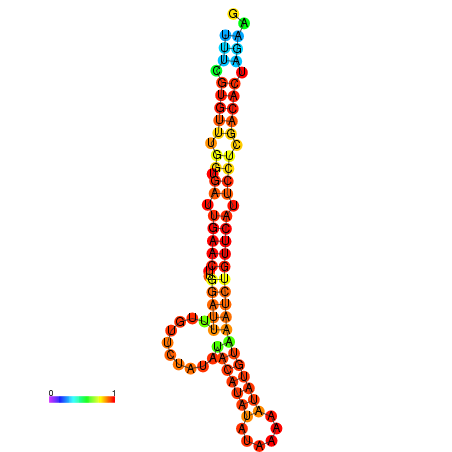 |
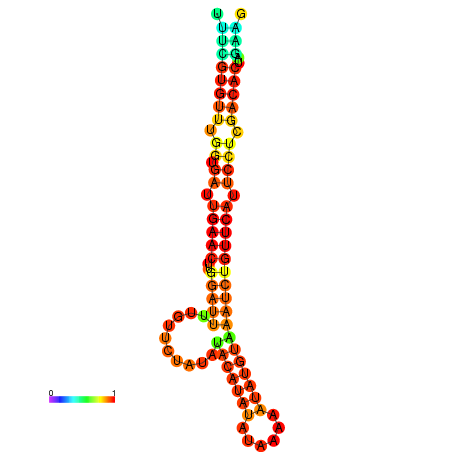 |
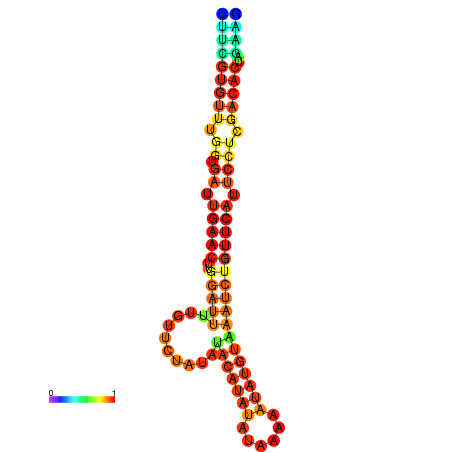 |
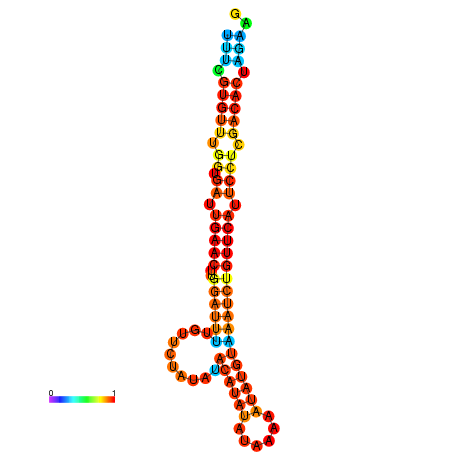 |
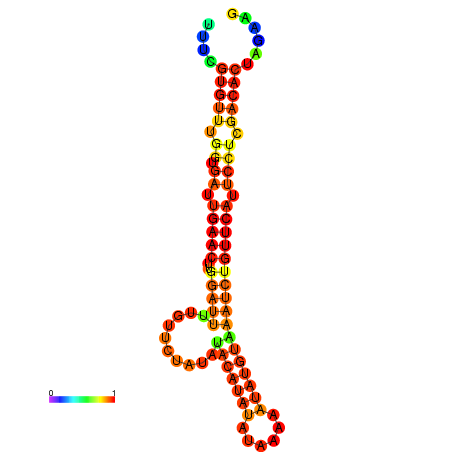 |
| dG=-12.7, p-value=0.009901 | dG=-12.7, p-value=0.009901 | dG=-12.5, p-value=0.009901 | dG=-12.5, p-value=0.009901 | dG=-12.4, p-value=0.009901 |
|---|---|---|---|---|
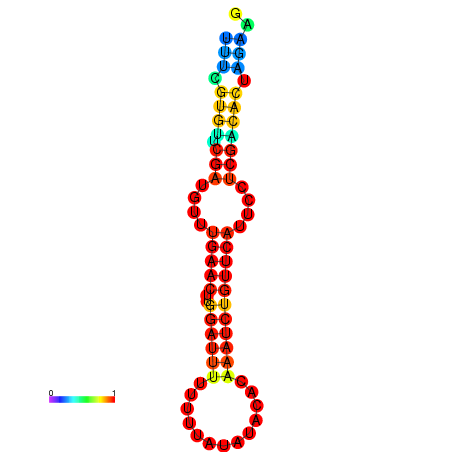 |
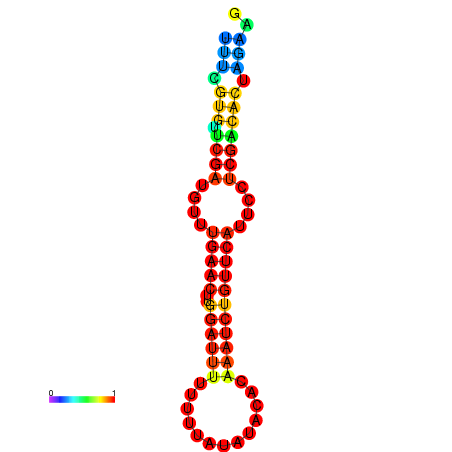 |
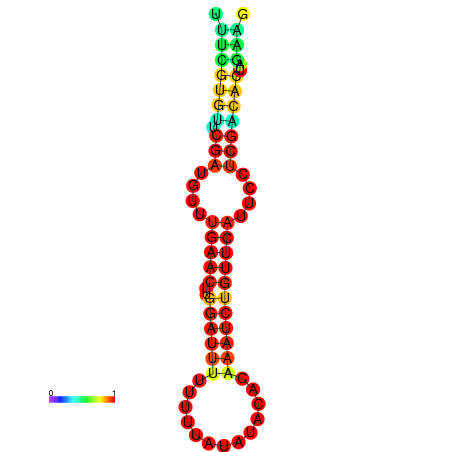 |
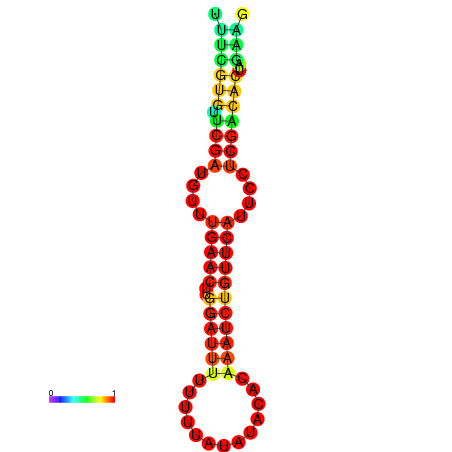 |
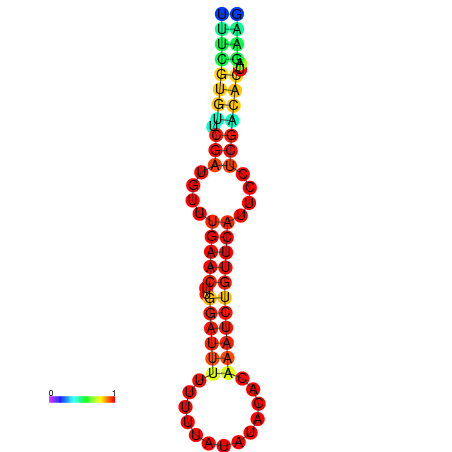 |
| dG=-14.8, p-value=0.009901 | dG=-14.8, p-value=0.009901 | dG=-14.8, p-value=0.009901 | dG=-14.8, p-value=0.009901 | dG=-14.2, p-value=0.009901 |
|---|---|---|---|---|
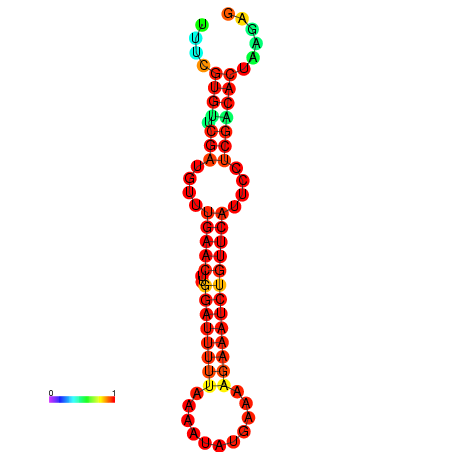 |
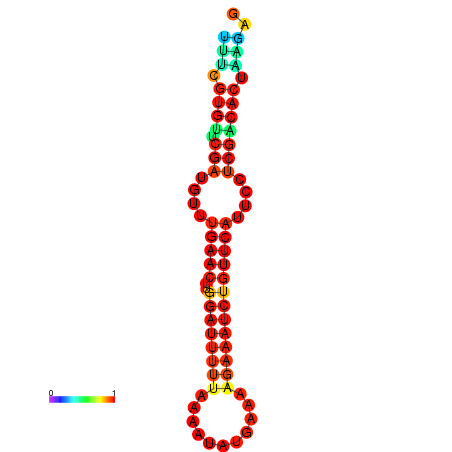 |
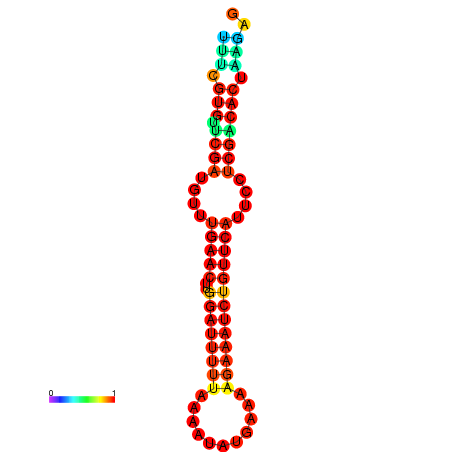 |
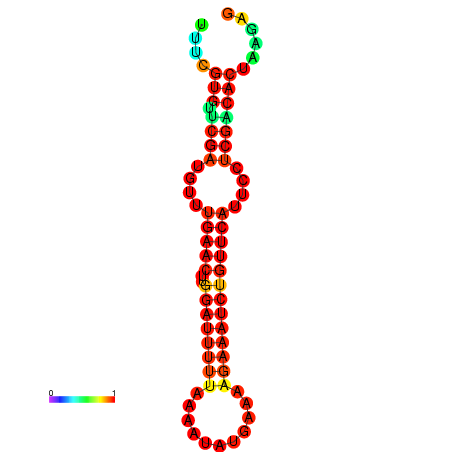 |
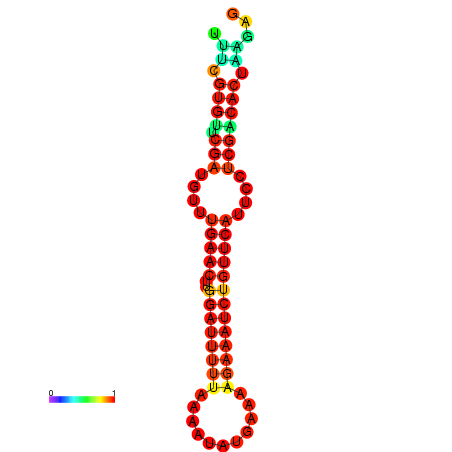 |
| dG=-11.6, p-value=0.009901 | dG=-11.0, p-value=0.009901 | dG=-10.6, p-value=0.009901 |
|---|---|---|
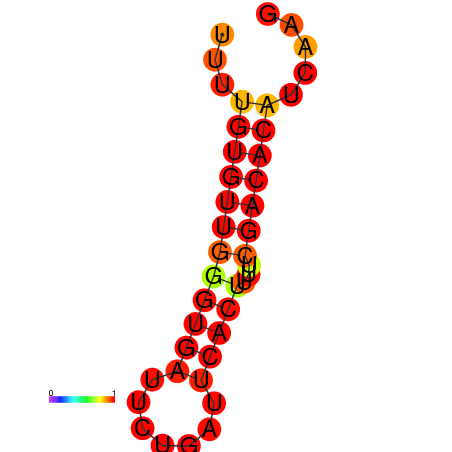 |
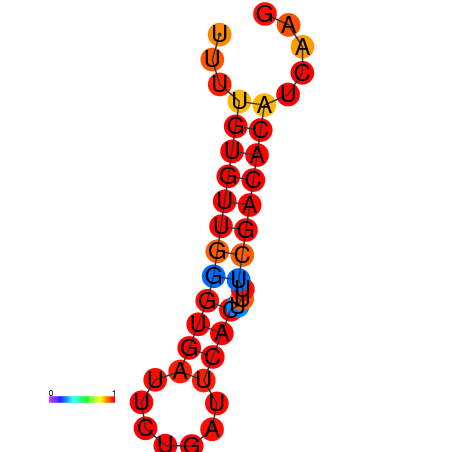 |
 |
Generated: 03/07/2013 at 07:17 PM