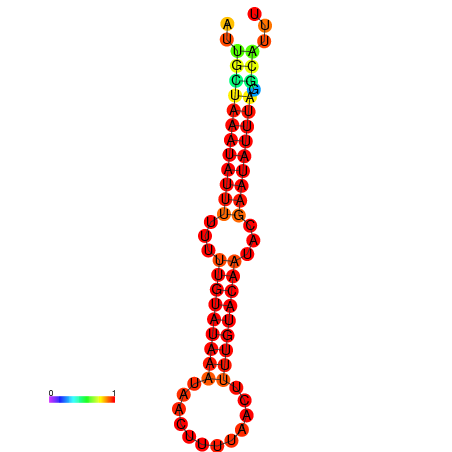
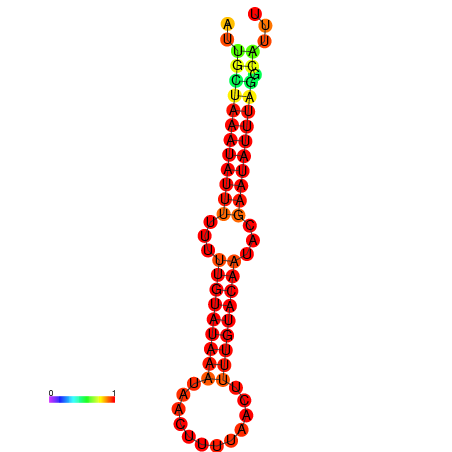
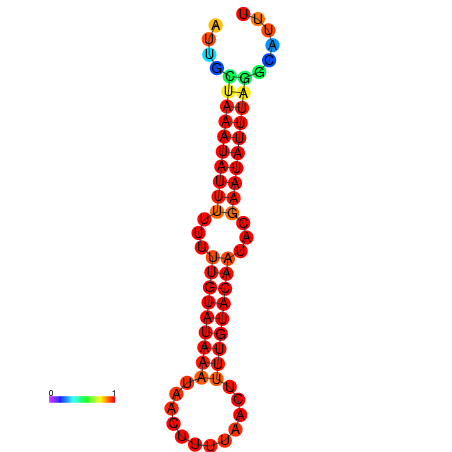
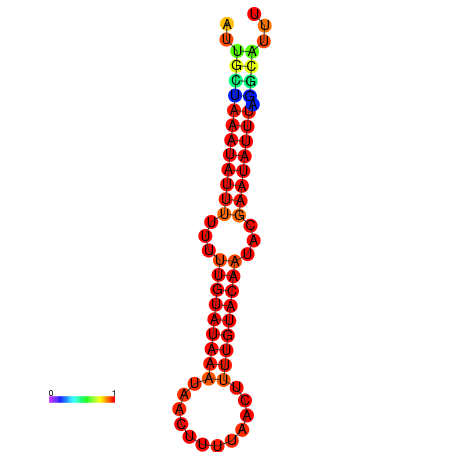
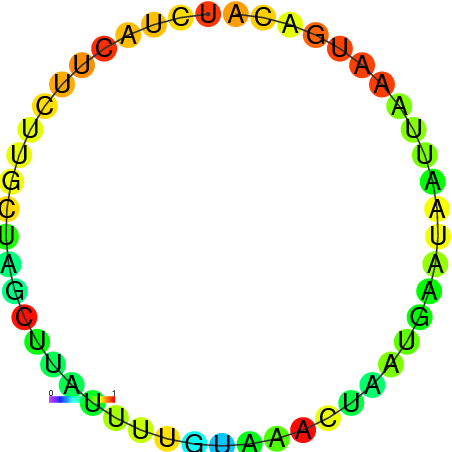
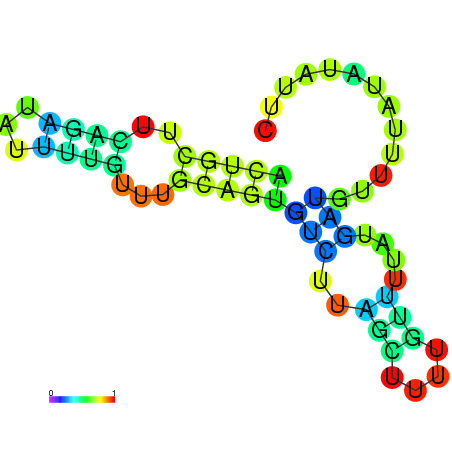
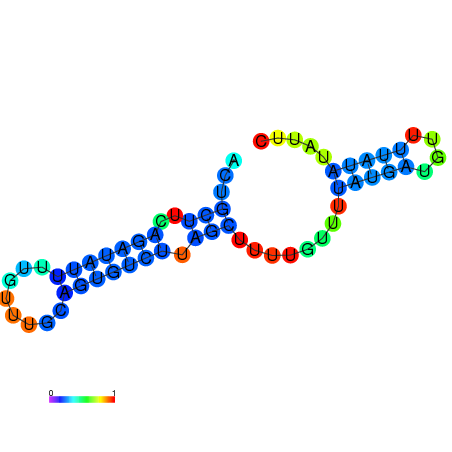
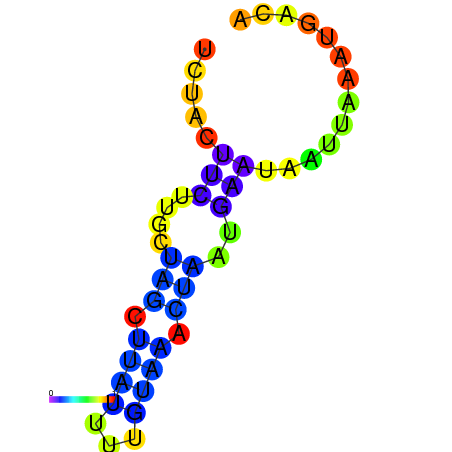| dm3 |
chrX:19445170-19445290 + |
TGCAAATAGCTAAAGAGAATGATAGGGAA----ATTGCT--AAATATTTTTTTTGTATAAATAACTTTT---AACTTTTGTACAATA------CG-AATATTTAGGCATTTCTCAAATCA------------------------------AA------------------------GATTTGCAAGCC-----------------AAA |
| droSim1 |
chrX:15010925-15011057 + |
ATAAAAAAGCTGAAGAGAATGATAGTG-----AATTGCT--AAATATTTTCTTTGTATAAATGACTTTT---AACATTTGTACAATA------TT-AATATTTAAACATTTCTCGAATCA------------------------------AAGTA-----------TGCCACATCAGCTTTGCAAGCC-----------------AAA |
| droSec1 |
super_8:1727594-1727726 + |
ATAAAAAAGCTGAAAAGAATGATAGTG-----AATTGCT--AAATATTTTCTTTGTATAAATAACTTTT---AATATTTGTACAATA------TT-AATATTTAAACATTTCTCGAATCA------------------------------AAGTA-----------TGCGACATCAGCTTTGCAAGCC-----------------AAA |
| droYak2 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droEre2 |
scaffold_4690:9681872-9682038 + |
-TGAAATAGCTGAATATAATGCTAGTGAA----ATTGTT--AAATATTTACTTTGTATAAATGACCTTCATAAACATTTATACAATT------TCAAATATTTTGCCATTTCTCAATCCAATTCAATTTTGCCACACAGAAGTGTCCAAAAAGTA-----------TGCTACATCATATTTGCAATCC-----------------AGA |
| droAna3 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dp4 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
scaffold_181096:7946002-7946145 + |
CTTAAATATTCGAAACGTGTACTAAGA-----TTCTACT--TCTTGCT------------------------AGCTTATTTTG--TAAACTAATG-AATAATTAAATGACAATGAAAACG------------------------------AAGTAAATTAGTTAATTAAGACAGCTGCTTAACATATAAAATAATTGAAAATATTCAA |
| droVir3 |
scaffold_13042:918660-918775 - |
CTGAAAGAACTATAGATATTATTAATAATAGGAACTGCTTCAGATATTTTGTTTGCAGT-----GTCTT---AGCTTTTGTTT--TA------TG-ATGTTTTATATATTCCGAATATTA------------------------------AA------------------------GCTTCAAA---------------------AAA |
| droMoj3 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |