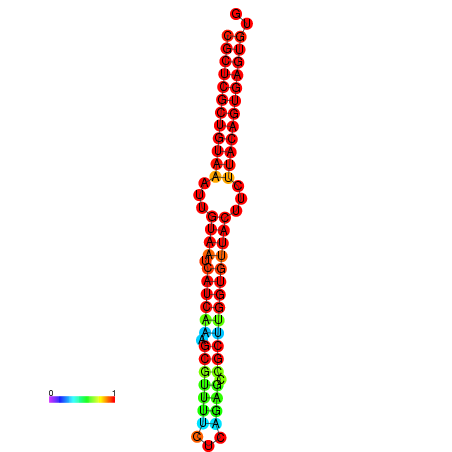
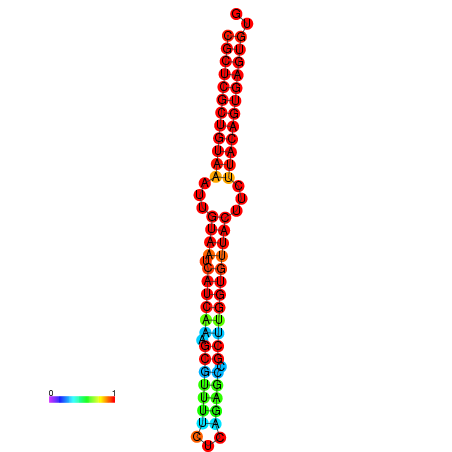
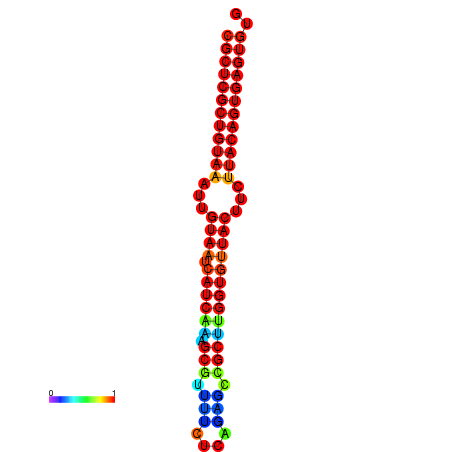
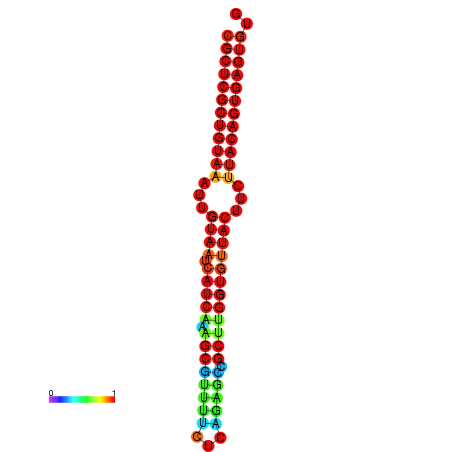
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:12956090-12956215 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droSim1 | zec46h04.g1:44-170 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droSec1 | super_21:701218-701343 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droYak2 | chrX:7229770-7229895 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAAAC--- |
| droEre2 | scaffold_4690:13914734-13914859 - | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCCCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droAna3 | scaffold_13117:3891784-3891909 + | TGCATGTGAGAAAA--------TTCCGTGGCTGGCAT--CGTTCACTGTAAATTGTAACCATCAAAGCGTT-TTTT----ATCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGAGCCAGCTG--------------------------GAGCACATATTAAAAA----- |
| dp4 | chrXL_group3a:1391638-1391748 - | AGCATGTGAGAGAGCGGCTCCTTTCCCTGGACCCCAT--CGCTCGCTGTAAATTGTAATGGCCAAAGCTTT-TTTG----TTGGTAGCAGCTTGGTGTTACTTCTTACAGTGAGTGTG------------------------------------------------------ |
| droPer1 | super_52:413307-413417 + | AGCATGTGAGAGAGCGGCTCCTTTCCCTGGACCCCAT--CGCTCGCTGTAAATTGTAATGGCCAAAGCTTT-TTTG----TTGGTAGCAGCTTGGTGTTACTTCTTACAGTGAGTGTG------------------------------------------------------ |
| droWil1 | scaffold_181096:6691507-6691601 - | AC-----------A--------TTAAACGGCCCAATA--CGCTCGCTGTAAATTGTAGCAATCAAAGCGTTGCCTTTTGCCTC---ACAGCTTGGTGTTACTTCTTACAGTGAGTGTG------------------------------------------------------ |
| droVir3 | scaffold_12928:4279754-4279849 - | ------------------------------------------CCGCTGTAAAATGTAATCATCAGAGCGTC-CT------CTC--AACCGCTTGGTGTTACTTCTTACAGTGAGTGCGACGGCCC--------------------------AAGGTCCGACCGTAAATCATA |
| droMoj3 | scaffold_6328:350121-350240 + | ---------------------------------------CGACCGCTGTAAATTGTAATCATCAAAGCGTC-TG------CTA--AATCGCTTGGTGTTACTTCTTACAGTGAGCGCGGCGCTCGAAACTACAGTGAAACTAGCATAAAGTGAACAAGGAAAGGAAA----- |
| droGri2 | scaffold_14853:994421-994540 - | CACG--------AA--------CT---TGAATGACGTCACGCCCGCTGTAATATGTAATCATCAAAGCGTG-TATT----TT---ACTCGCTTGGTGTTACTTCTTACAGTGAGTGCGACGGCCACAC-----------------------GTCCAGCGGAGGAGAGTG--- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:12956090-12956215 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droSim1 | zec46h04.g1:44-170 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droSec1 | super_21:701218-701343 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droYak2 | chrX:7229770-7229895 + | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCTCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAAAC--- |
| droEre2 | scaffold_4690:13914734-13914859 - | TGCATGTGAGAGAA--------TTCCGTGGCTGGCAT--CGCCCGCTGTAAATTGTAATCATCAAAGCGTT-TT------CTCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGTGCCAGTCC--------------------------GTACACAGAAAGAAAACC--- |
| droAna3 | scaffold_13117:3891784-3891909 + | TGCATGTGAGAAAA--------TTCCGTGGCTGGCAT--CGTTCACTGTAAATTGTAACCATCAAAGCGTT-TTTT----ATCAGAGCCGCTTGGTGTTACTTCTTACAGTGAGTGAGCCAGCTG--------------------------GAGCACATATTAAAAA----- |
| dp4 | chrXL_group3a:1391638-1391748 - | AGCATGTGAGAGAGCGGCTCCTTTCCCTGGACCCCAT--CGCTCGCTGTAAATTGTAATGGCCAAAGCTTT-TTTG----TTGGTAGCAGCTTGGTGTTACTTCTTACAGTGAGTGTG------------------------------------------------------ |
| droPer1 | super_52:413307-413417 + | AGCATGTGAGAGAGCGGCTCCTTTCCCTGGACCCCAT--CGCTCGCTGTAAATTGTAATGGCCAAAGCTTT-TTTG----TTGGTAGCAGCTTGGTGTTACTTCTTACAGTGAGTGTG------------------------------------------------------ |
| droWil1 | scaffold_181096:6691507-6691601 - | AC-----------A--------TTAAACGGCCCAATA--CGCTCGCTGTAAATTGTAGCAATCAAAGCGTTGCCTTTTGCCTC---ACAGCTTGGTGTTACTTCTTACAGTGAGTGTG------------------------------------------------------ |
| droVir3 | scaffold_12928:4279754-4279849 - | ------------------------------------------CCGCTGTAAAATGTAATCATCAGAGCGTC-CT------CTC--AACCGCTTGGTGTTACTTCTTACAGTGAGTGCGACGGCCC--------------------------AAGGTCCGACCGTAAATCATA |
| droMoj3 | scaffold_6328:350121-350240 + | ---------------------------------------CGACCGCTGTAAATTGTAATCATCAAAGCGTC-TG------CTA--AATCGCTTGGTGTTACTTCTTACAGTGAGCGCGGCGCTCGAAACTACAGTGAAACTAGCATAAAGTGAACAAGGAAAGGAAA----- |
| droGri2 | scaffold_14853:994421-994540 - | CACG--------AA--------CT---TGAATGACGTCACGCCCGCTGTAATATGTAATCATCAAAGCGTG-TATT----TT---ACTCGCTTGGTGTTACTTCTTACAGTGAGTGCGACGGCCACAC-----------------------GTCCAGCGGAGGAGAGTG--- |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
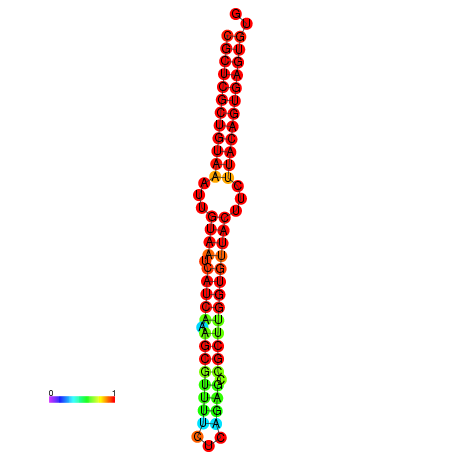
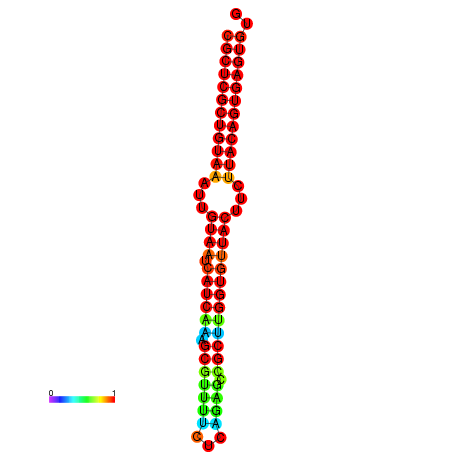 |
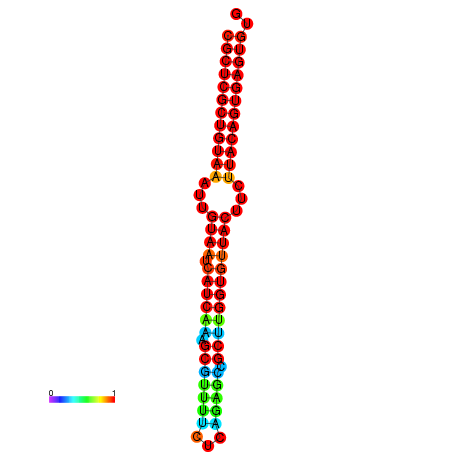 |
 |
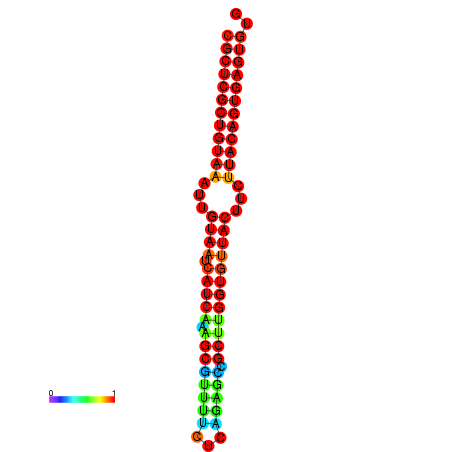 |
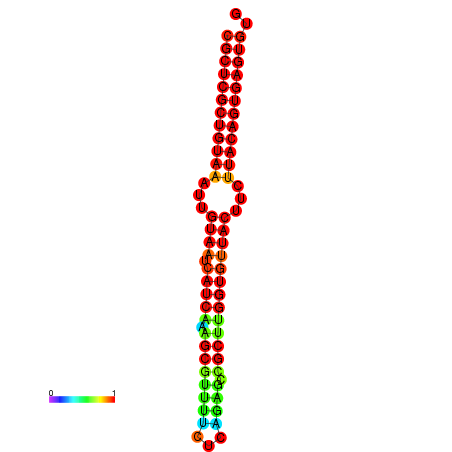 |
| dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
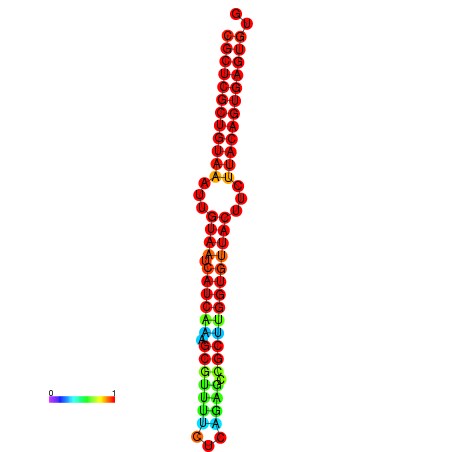 |
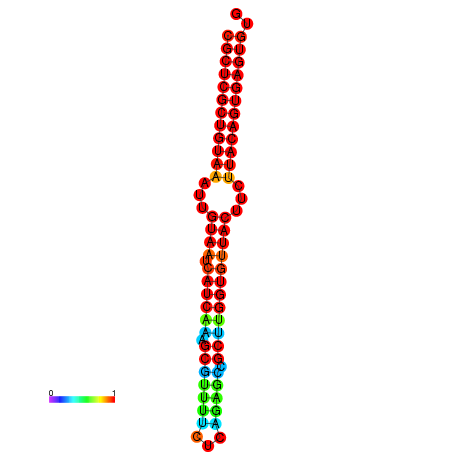 |
 |
 |
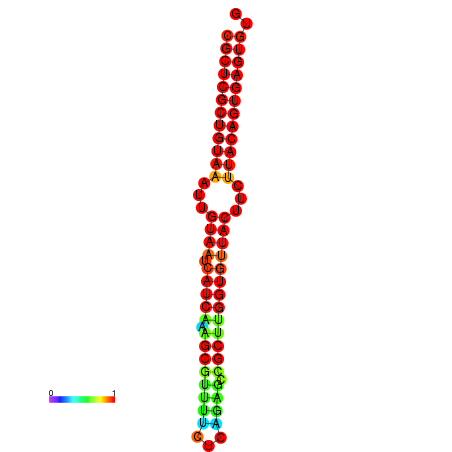 |
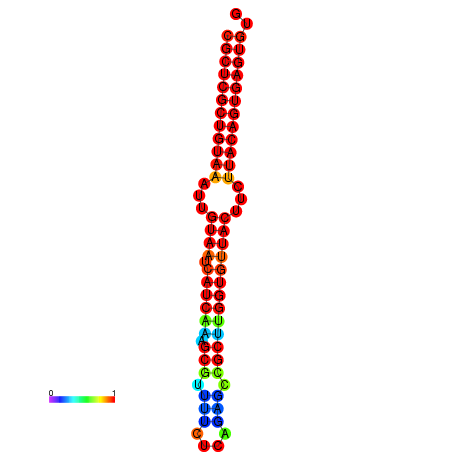
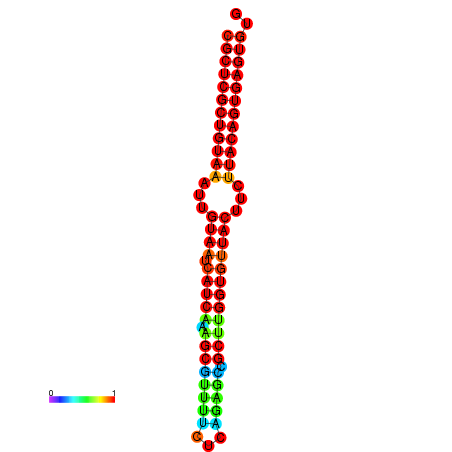
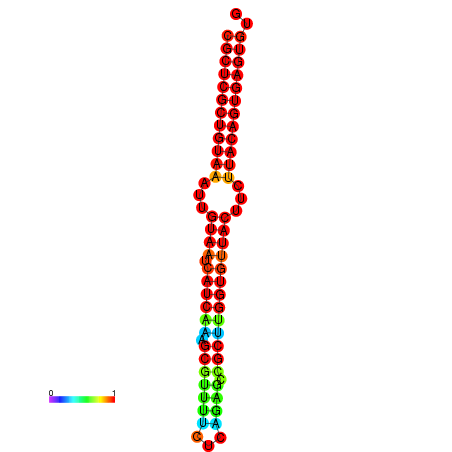
| dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
 |
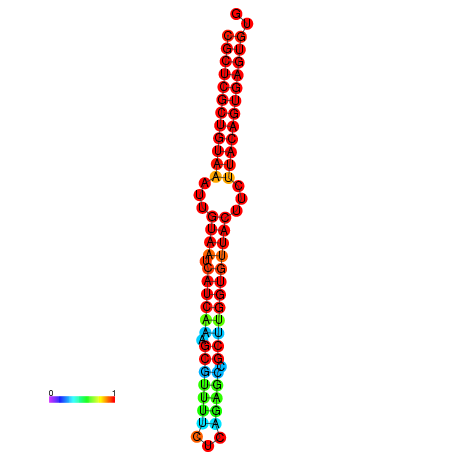 |
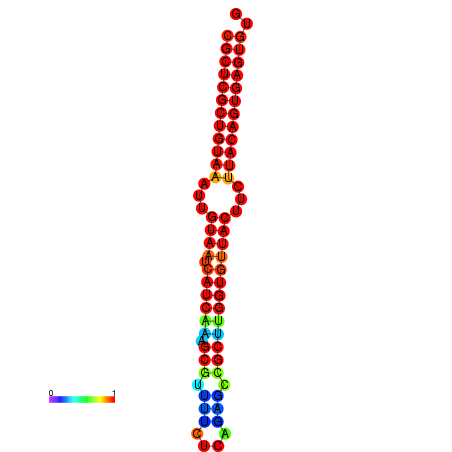 |
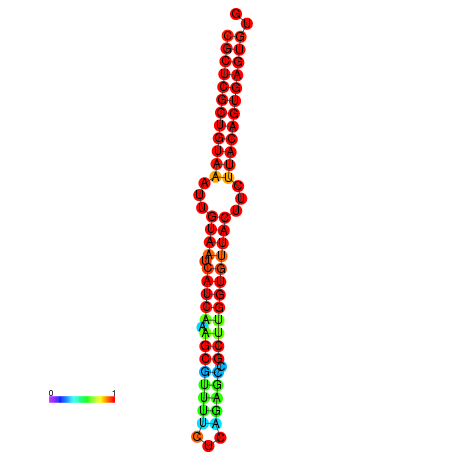 |
 |
| dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 |
|---|---|---|---|---|
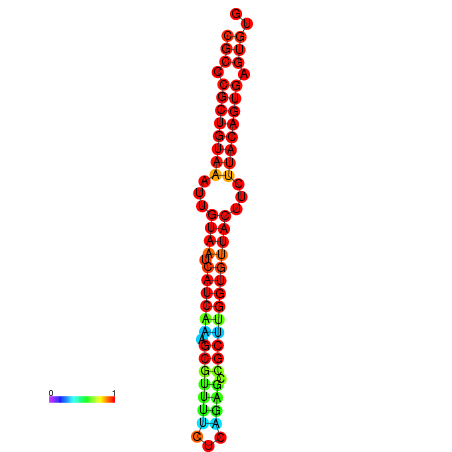 |
 |
 |
 |
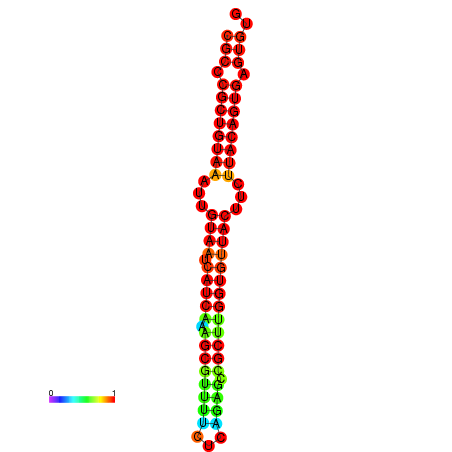 |
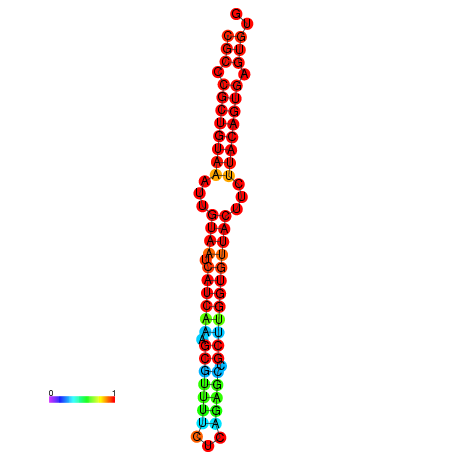
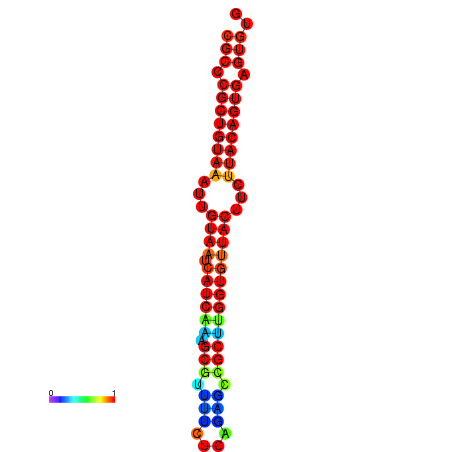
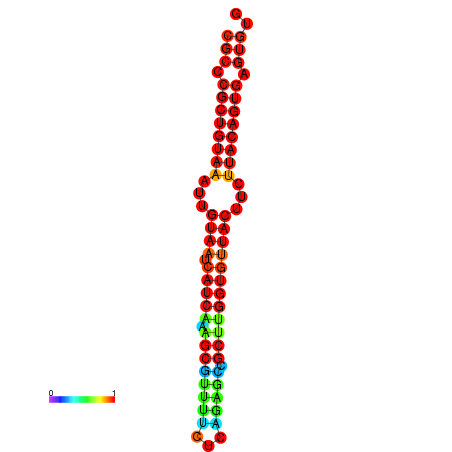
| dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 |
|---|---|---|---|---|
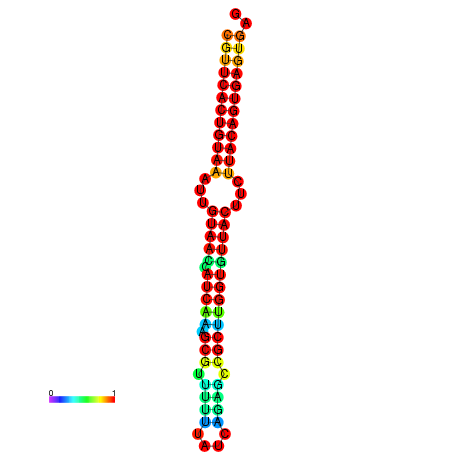 |
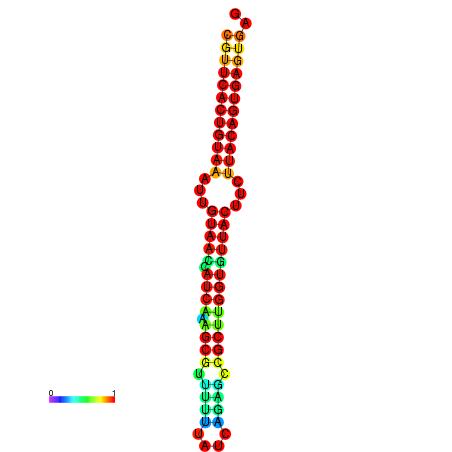 |
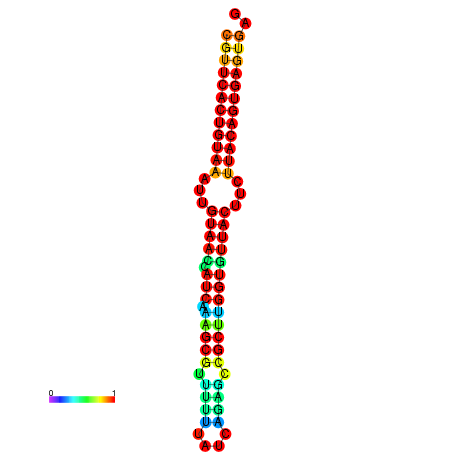 |
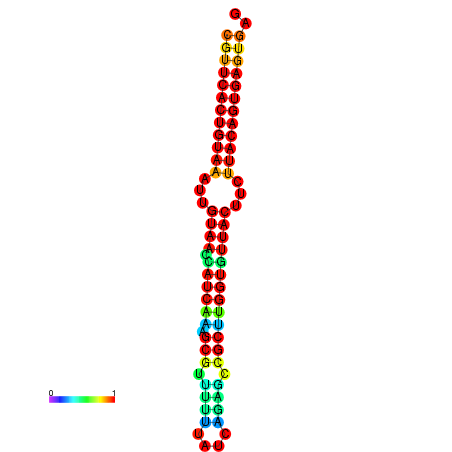 |
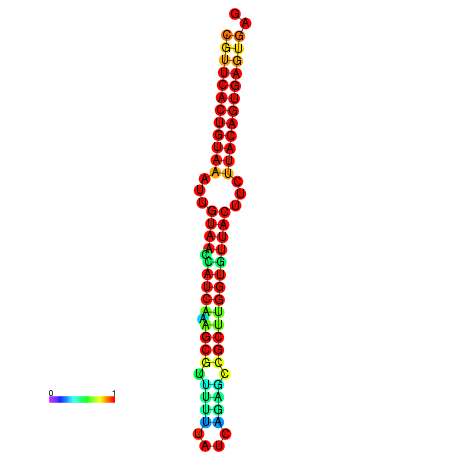 |
| dG=-28.5, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
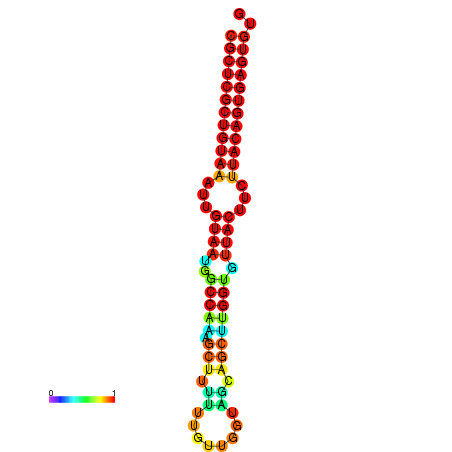 |
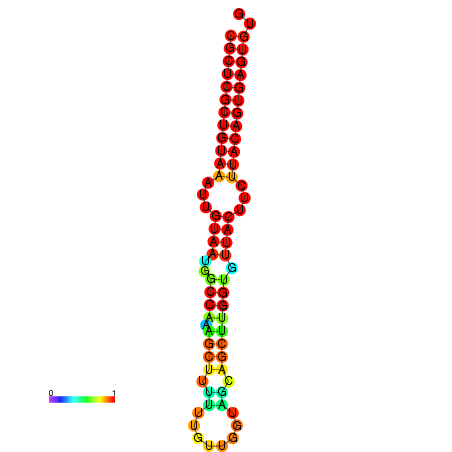 |
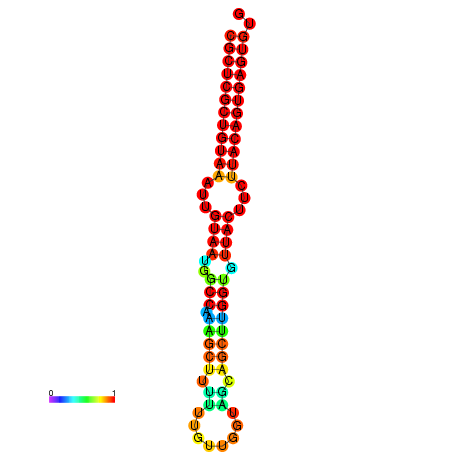 |
 |
 |
| dG=-28.5, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
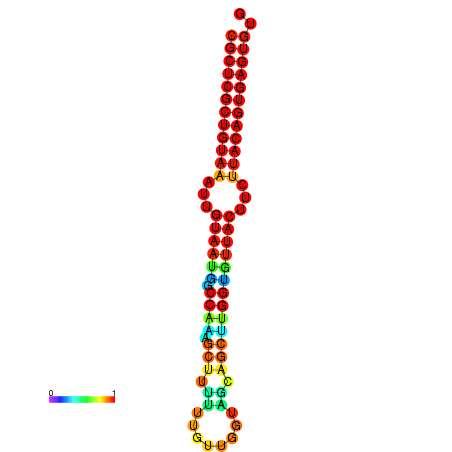 |
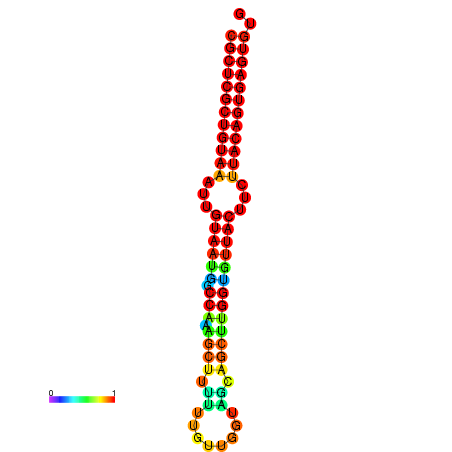 |
| dG=-28.8, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.8, p-value=0.009901 |
|---|---|---|---|---|
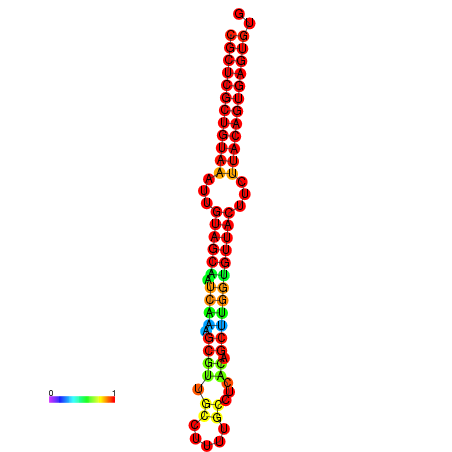 |
 |
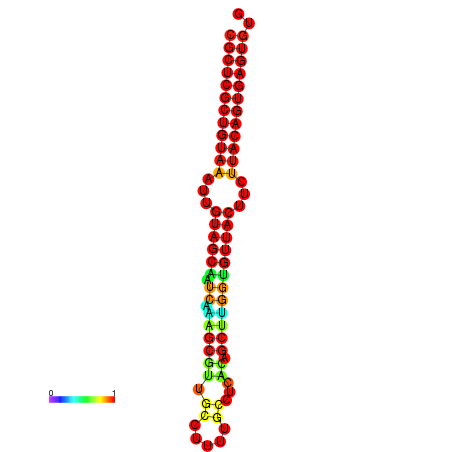 |
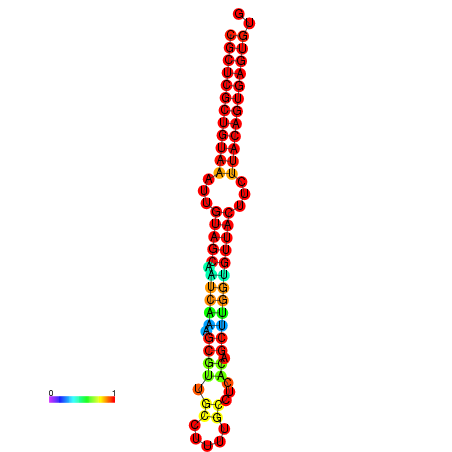 |
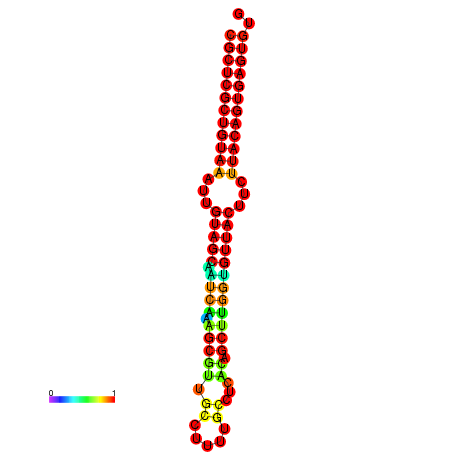 |
| dG=-20.8, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.5, p-value=0.009901 |
|---|---|---|
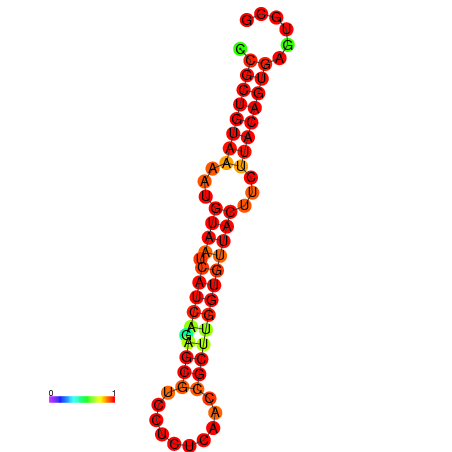 |
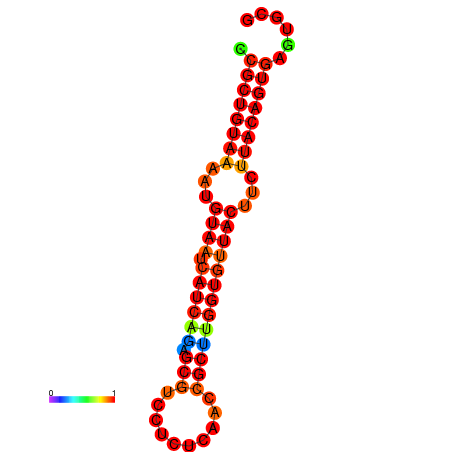 |
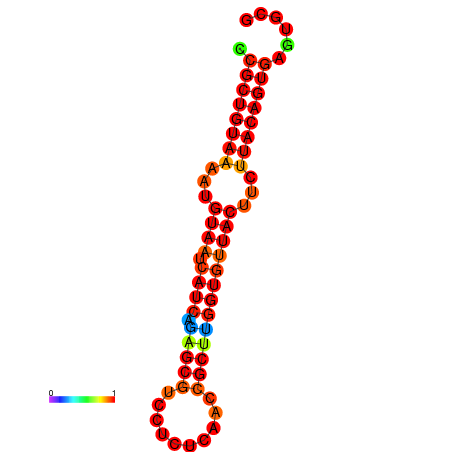 |
| dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|
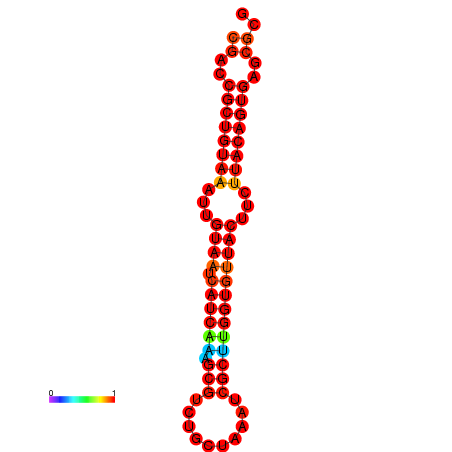 |
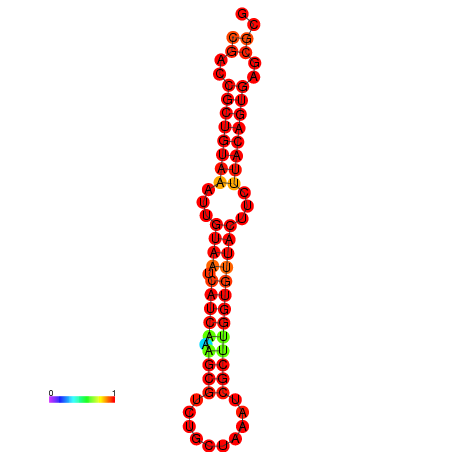 |
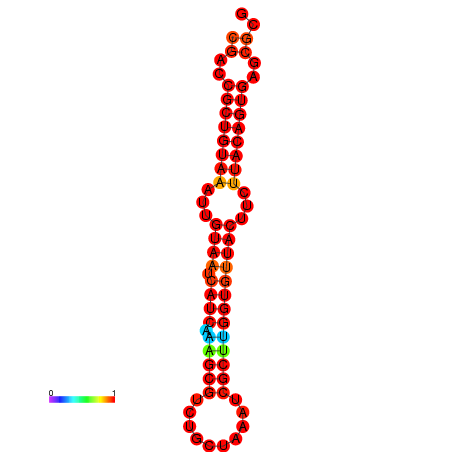 |
| dG=-27.4, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.6, p-value=0.009901 |
|---|---|---|---|---|
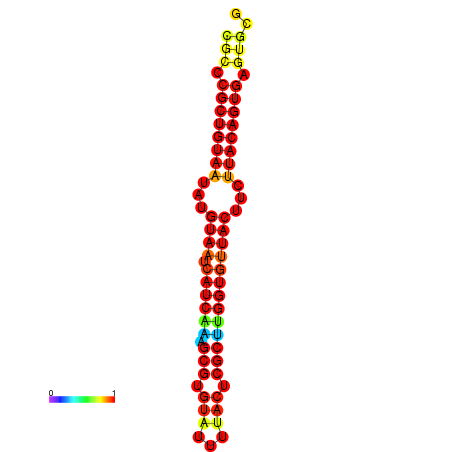 |
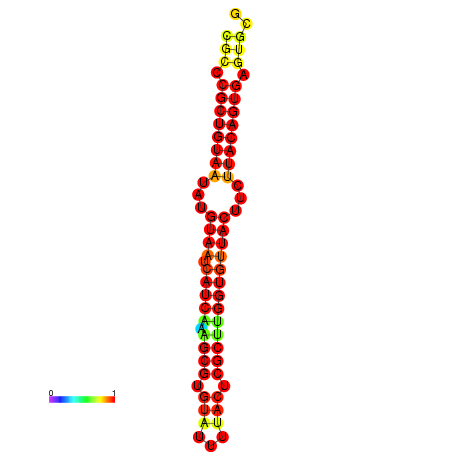 |
 |
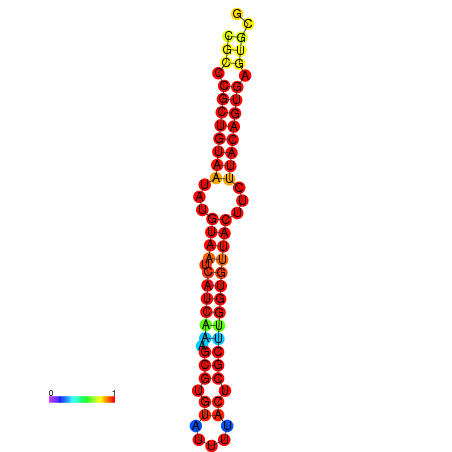 |
 |
Generated: 03/07/2013 at 07:16 PM