| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:18018375-18018498 + | TTCGG---------------TGAATCC--CCAAGTCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAC----TACTGC-----AATTCGAAA--T |
| droSim1 | chrU:15519348-15519471 + | TTCGG---------------GGAATCC--CCAAGTCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAC----TACTGC-----AATTCAAAA--T |
| droSec1 | super_8:326574-326685 + | TTCGA---------------GGAATCC--CCAAGTCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAC----TACT------------------- |
| droYak2 | chrX:16642309-16642432 + | TTCGG---------------GGTATCC--CCAAACCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTTGTGGGA--TGGTC----TACCGC-----AATTCGAAA--G |
| droEre2 | scaffold_4690:8346009-8346128 + | TTTGG---------------GAAGTTC--CCAAATCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAT----TACTGC---------CGAAA--G |
| droAna3 | scaffold_13335:1607974-1608101 - | TGTGT--TTATTTTTTTT--GGAA-------AATATTCTGTTTTACGTGCGAGTTCCACTAAGCAAGTTTTGAGAATTT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTCCGTGGGG--CAGAT----AATGTC-----AG--CAGTA--T |
| dp4 | chrXL_group3a:1977519-1977620 - | TTCCT---------------GGAAGCCATCTAGGAGCCTGTTCTACGTGCGAGTTCCACCATGCAAGTTTTGAGC--GTATATGAATCCAAAAACTTGACACGTTGAGCTCGTGCGTGG------------------------------------ |
| droPer1 | super_11:2701270-2701398 + | TTCCT---------------GGAAGCCATCTAGGAGCCTGTTCTACGTGCGAGTTCCACCATGCAAGTTTTGAGC--GTATATGAATCCAAAAACTTGACACGTTGAGCTCGTGCGTGGTG--CAGCCAAATCATCGC-----AGCGCATCC--A |
| droWil1 | scaffold_181096:12029390-12029503 + | TTTAT-----------------------------ATCCTGTCCTACGCATGAGTTCCACTAAGCAAGTTTTGGGCATAT-TTCAAAAACAAAAACTTGACACGTTGAGCTCGTCTGTGTGA--CAGAA----C-GAACAAC--AAATGAAGA--A |
| droVir3 | scaffold_12928:1264092-1264205 - | TTCAA---------------GAGCAGC--TCGCGCTTCTGCCCTACGCACGAGTTCCACTAATCAAGTTTTGAGCGGAT-TCGAAAGTCGAAAACTTGACACGTTGAGCTCGTCCGTGGGG--CGGCA----TTC---------------AA--G |
| droMoj3 | scaffold_6473:13724290-13724417 + | TCCTTATCCATAC-------CATATCC--ACATTTGGCTGTCCTACGAGCGAGTTCCACTAATCAAGTTTTGAGT--GT-TCGAAAATCAAAAACTTGACACGTTGAGCTCGTCTGTGTGG--CA--C----AAGTTC-----AA--CAAAAGCA |
| droGri2 | scaffold_15203:8445730-8445869 + | TTCGGT-TTGTTCTTTTACAGCAGTGCATCCGGGTGTCTGCCACACTTACGAGTTCCACTATTCAAGTTTTGAGC--GTATTTGAAATCAAAAACTTGACACGTTGAGCTCGTGTGTGCGTTGCGG------AAA-TCAACTTA---CTCAA--A |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:18018375-18018498 + | TTCGG---------------TGAATCC--CCAAGTCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAC----TACTGC-----AATTCGAAA--T |
| droSim1 | chrU:15519348-15519471 + | TTCGG---------------GGAATCC--CCAAGTCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAC----TACTGC-----AATTCAAAA--T |
| droSec1 | super_8:326574-326685 + | TTCGA---------------GGAATCC--CCAAGTCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAC----TACT------------------- |
| droYak2 | chrX:16642309-16642432 + | TTCGG---------------GGTATCC--CCAAACCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTTGTGGGA--TGGTC----TACCGC-----AATTCGAAA--G |
| droEre2 | scaffold_4690:8346009-8346128 + | TTTGG---------------GAAGTTC--CCAAATCTCTGTCCTACGTCCGAGTTCCACTAAGCAAGTTTTGAGATCGT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTTCGTGGGA--TGGAT----TACTGC---------CGAAA--G |
| droAna3 | scaffold_13335:1607974-1608101 - | TGTGT--TTATTTTTTTT--GGAA-------AATATTCTGTTTTACGTGCGAGTTCCACTAAGCAAGTTTTGAGAATTT-TTTAAAAACAAAAACTTGACACGTTGAGCTCGTCCGTGGGG--CAGAT----AATGTC-----AG--CAGTA--T |
| dp4 | chrXL_group3a:1977519-1977620 - | TTCCT---------------GGAAGCCATCTAGGAGCCTGTTCTACGTGCGAGTTCCACCATGCAAGTTTTGAGC--GTATATGAATCCAAAAACTTGACACGTTGAGCTCGTGCGTGG------------------------------------ |
| droPer1 | super_11:2701270-2701398 + | TTCCT---------------GGAAGCCATCTAGGAGCCTGTTCTACGTGCGAGTTCCACCATGCAAGTTTTGAGC--GTATATGAATCCAAAAACTTGACACGTTGAGCTCGTGCGTGGTG--CAGCCAAATCATCGC-----AGCGCATCC--A |
| droWil1 | scaffold_181096:12029390-12029503 + | TTTAT-----------------------------ATCCTGTCCTACGCATGAGTTCCACTAAGCAAGTTTTGGGCATAT-TTCAAAAACAAAAACTTGACACGTTGAGCTCGTCTGTGTGA--CAGAA----C-GAACAAC--AAATGAAGA--A |
| droVir3 | scaffold_12928:1264092-1264205 - | TTCAA---------------GAGCAGC--TCGCGCTTCTGCCCTACGCACGAGTTCCACTAATCAAGTTTTGAGCGGAT-TCGAAAGTCGAAAACTTGACACGTTGAGCTCGTCCGTGGGG--CGGCA----TTC---------------AA--G |
| droMoj3 | scaffold_6473:13724290-13724417 + | TCCTTATCCATAC-------CATATCC--ACATTTGGCTGTCCTACGAGCGAGTTCCACTAATCAAGTTTTGAGT--GT-TCGAAAATCAAAAACTTGACACGTTGAGCTCGTCTGTGTGG--CA--C----AAGTTC-----AA--CAAAAGCA |
| droGri2 | scaffold_15203:8445730-8445869 + | TTCGGT-TTGTTCTTTTACAGCAGTGCATCCGGGTGTCTGCCACACTTACGAGTTCCACTATTCAAGTTTTGAGC--GTATTTGAAATCAAAAACTTGACACGTTGAGCTCGTGTGTGCGTTGCGG------AAA-TCAACTTA---CTCAA--A |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-19.7, p-value=0.009901 |
|---|
 |
| dG=-19.7, p-value=0.009901 |
|---|
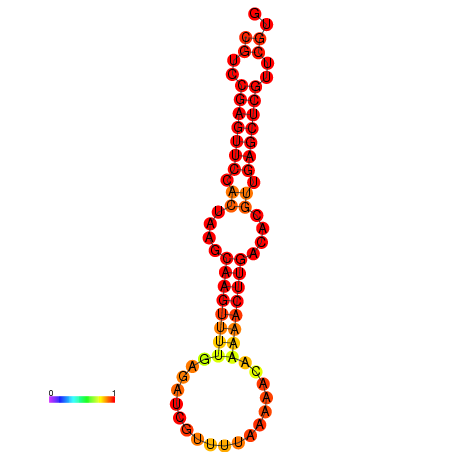
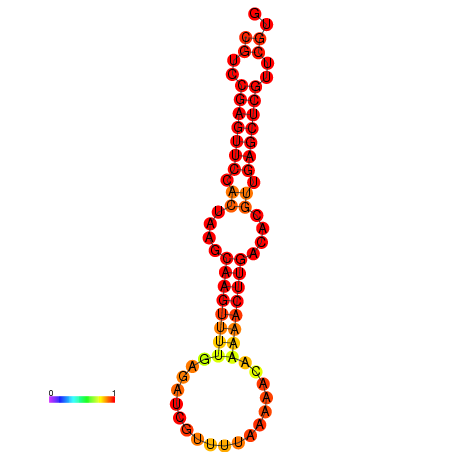 |
| dG=-19.7, p-value=0.009901 |
|---|
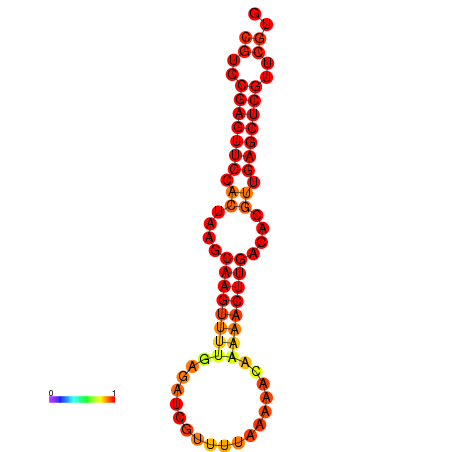 |
| dG=-17.1, p-value=0.009901 | dG=-16.7, p-value=0.009901 |
|---|---|
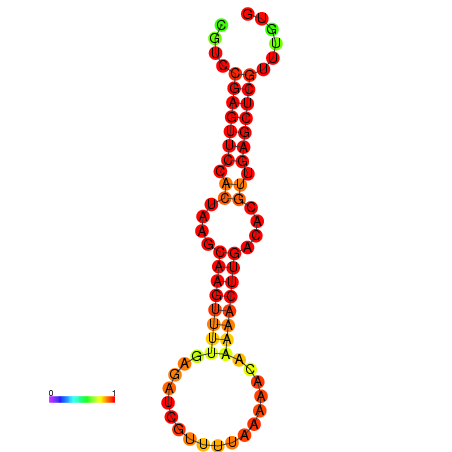 |
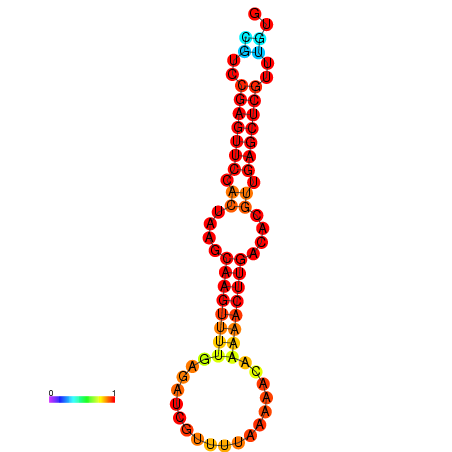 |
| dG=-19.7, p-value=0.009901 |
|---|
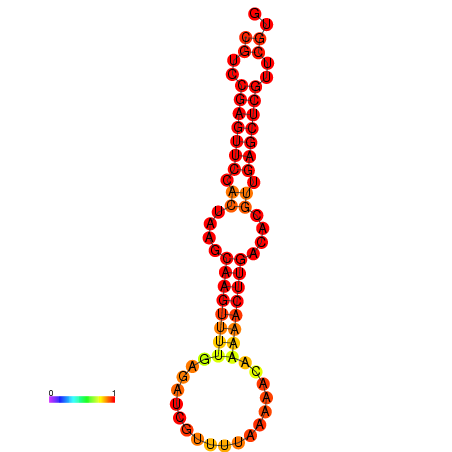 |
| dG=-21.2, p-value=0.009901 |
|---|
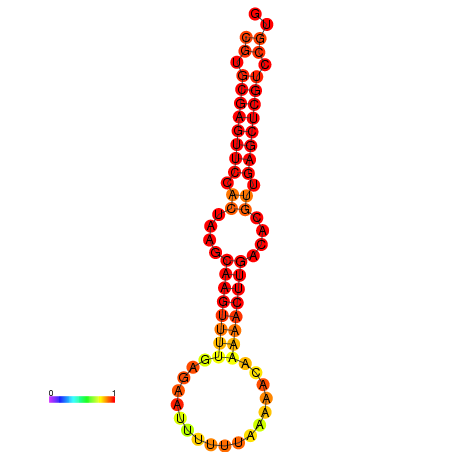 |
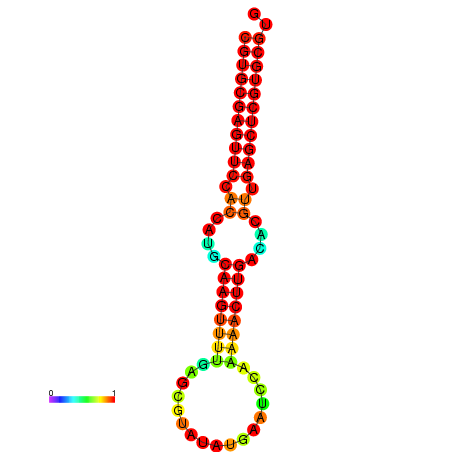
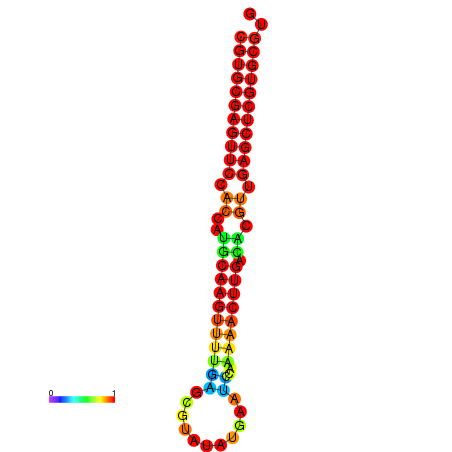
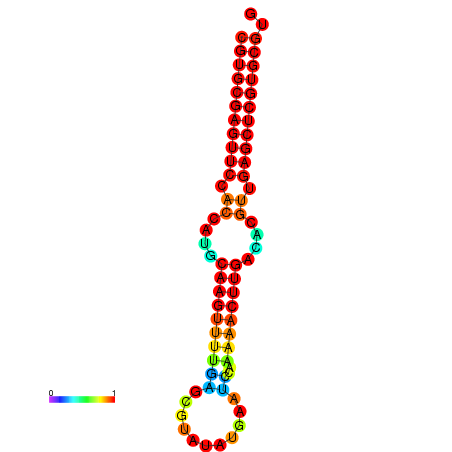
| dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
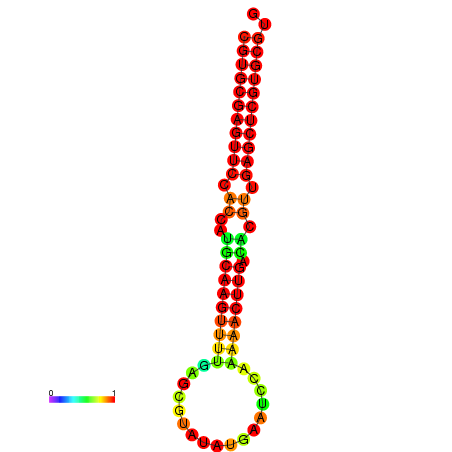 |
 |
 |
 |
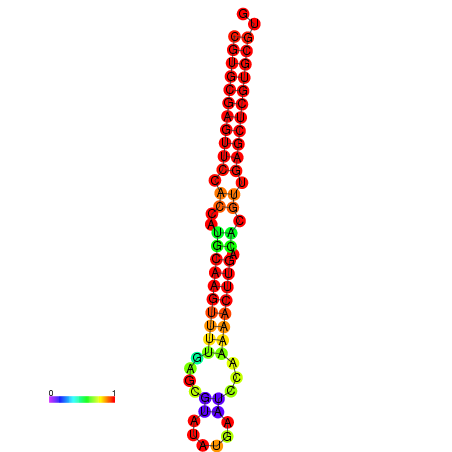 |
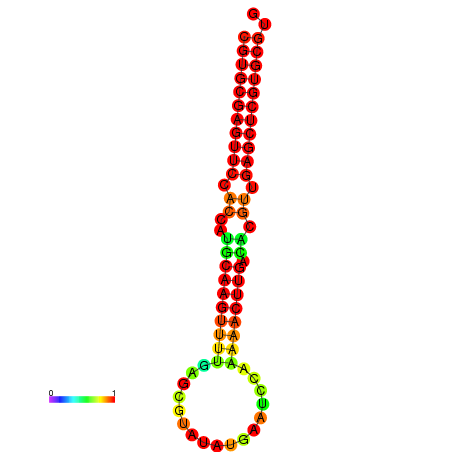
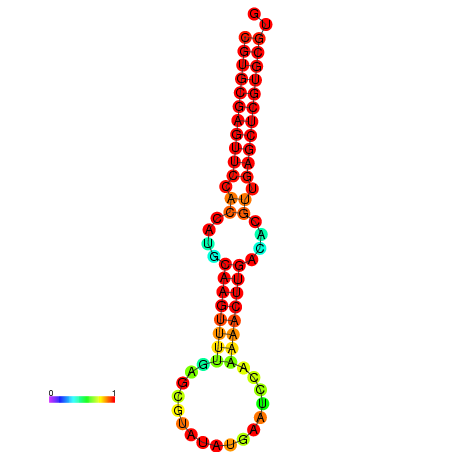
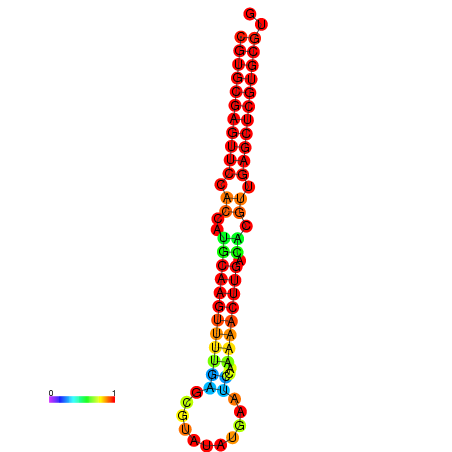
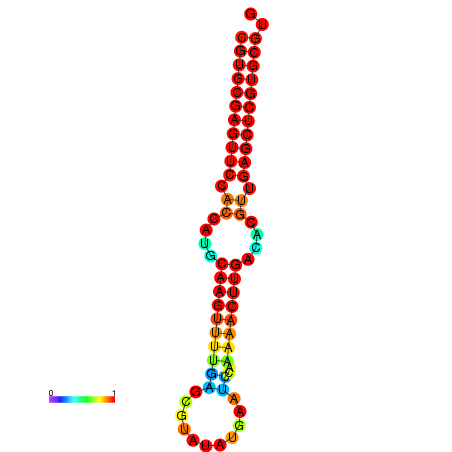
| dG=-24.8, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-17.5, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.3, p-value=0.009901 | dG=-17.0, p-value=0.009901 | dG=-17.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
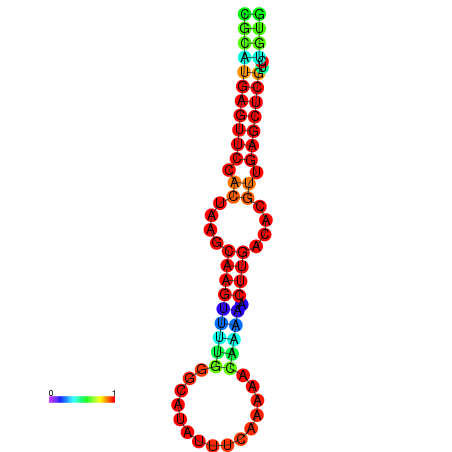 |
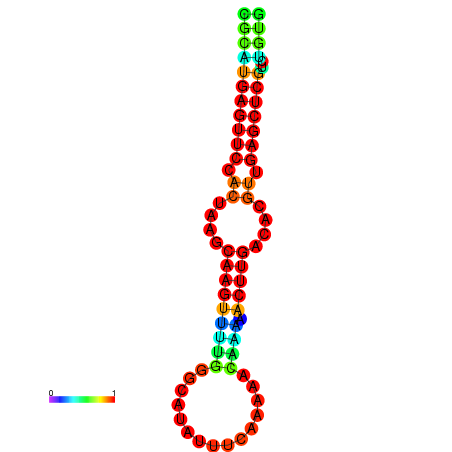 |
| dG=-24.8, p-value=0.009901 |
|---|
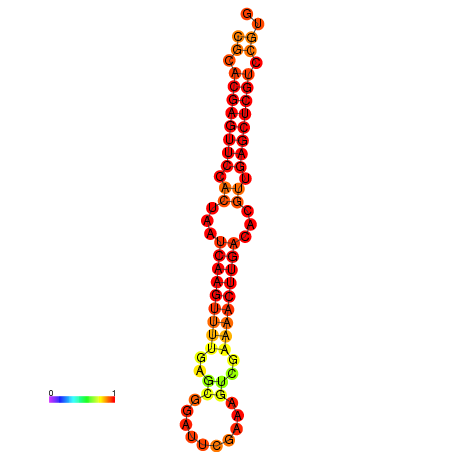 |
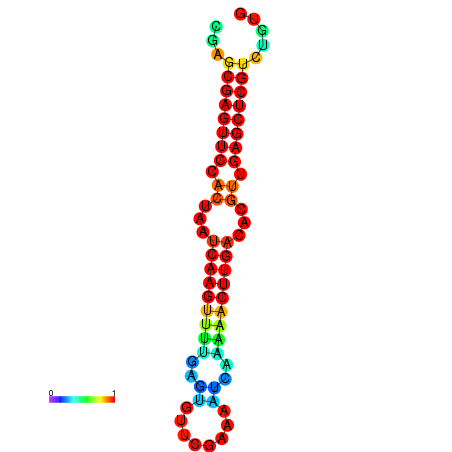
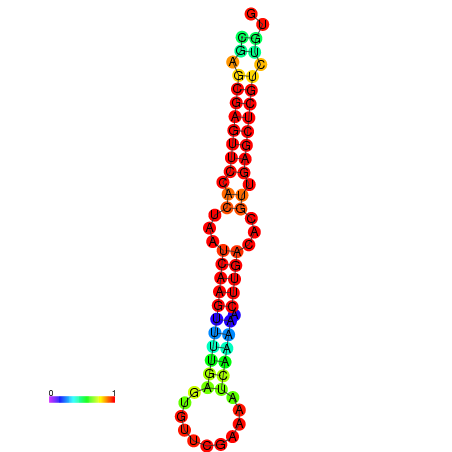
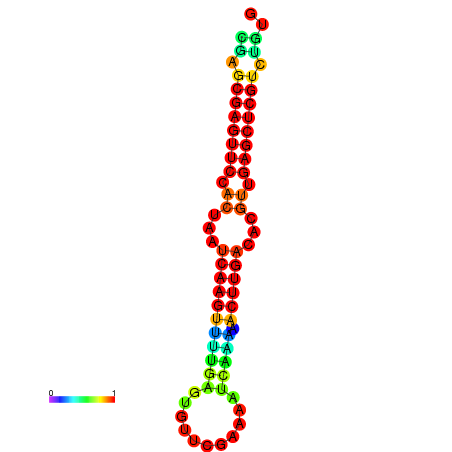
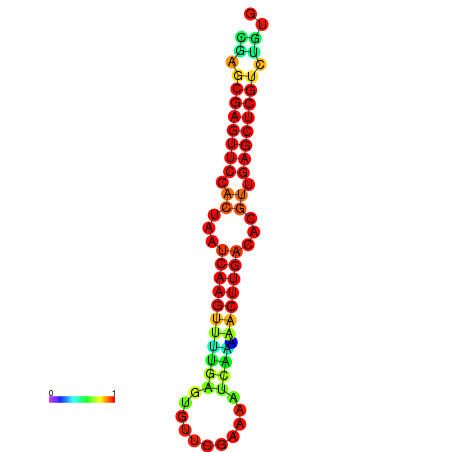
| dG=-22.0, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.6, p-value=0.009901 |
|---|---|---|---|---|
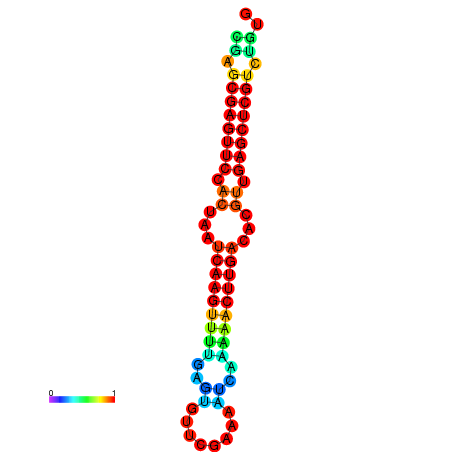 |
 |
 |
 |
 |
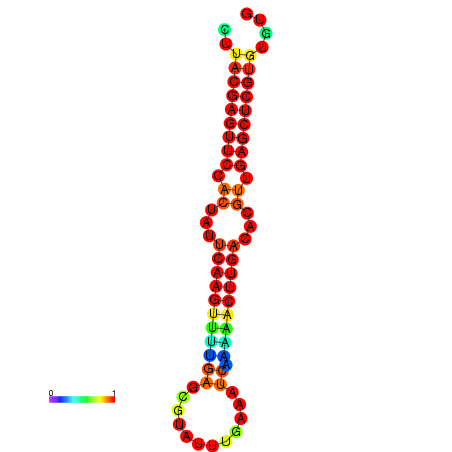
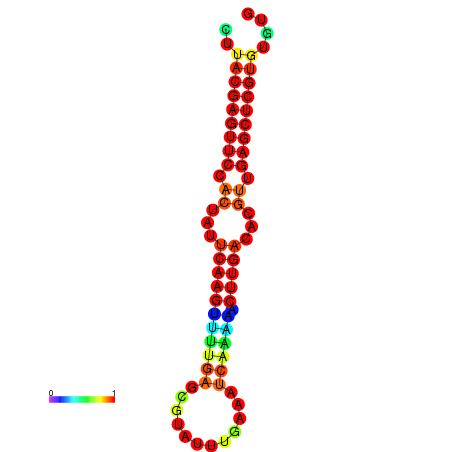
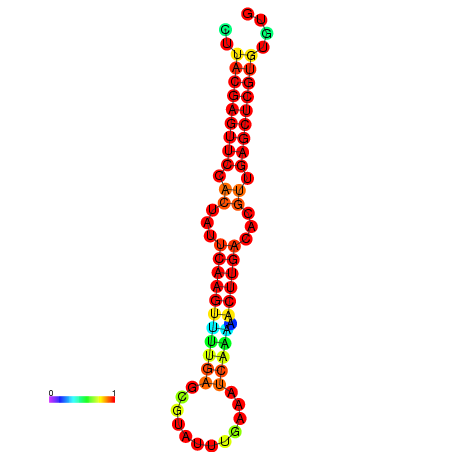
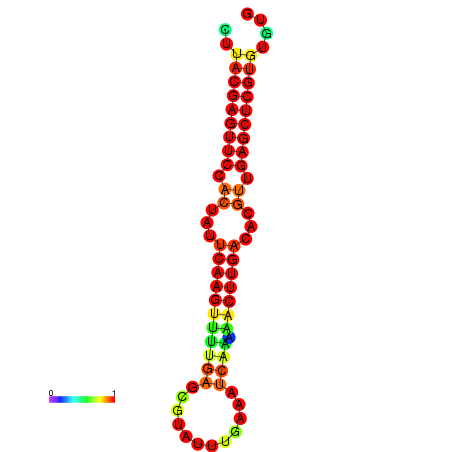
| dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 07:14 PM