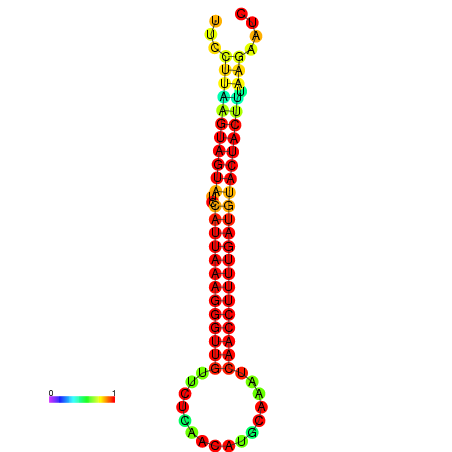
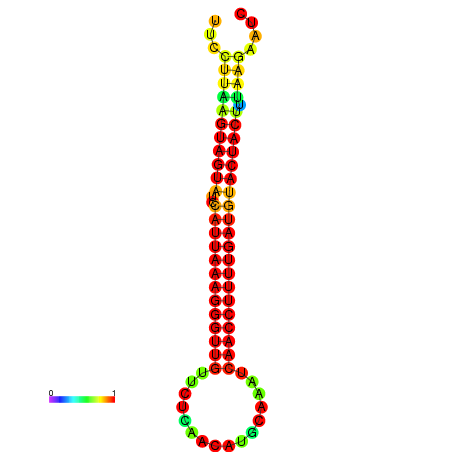
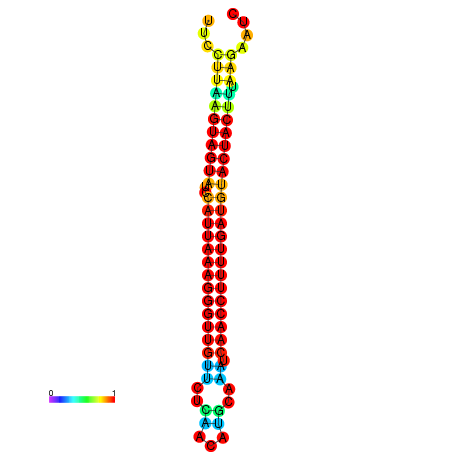
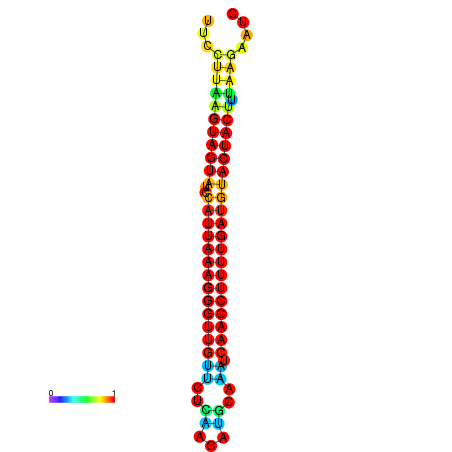
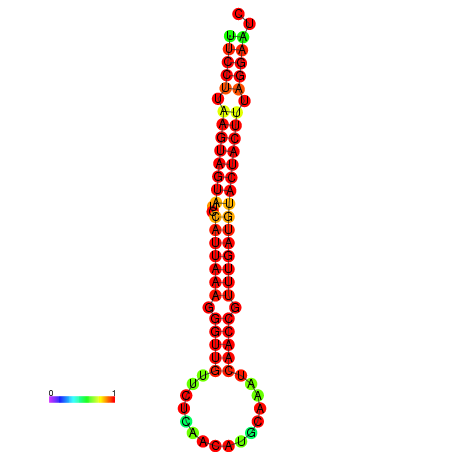
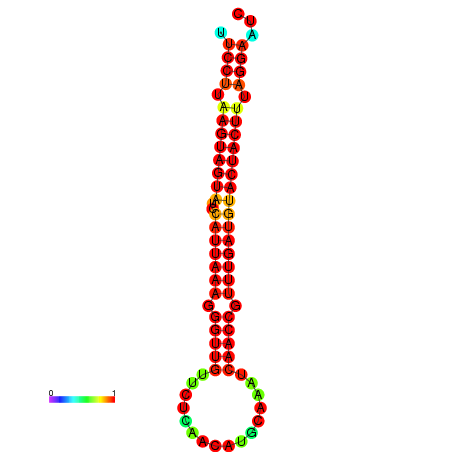
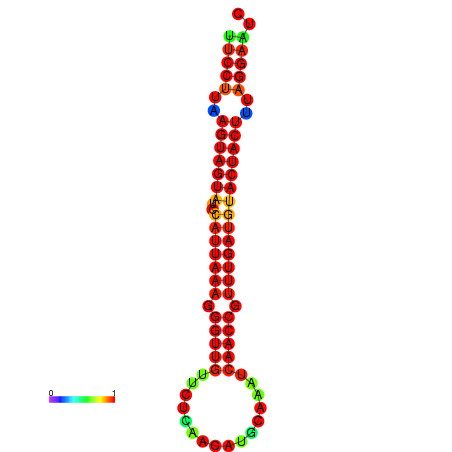
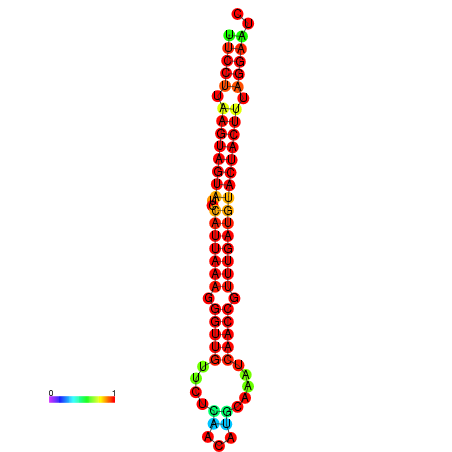
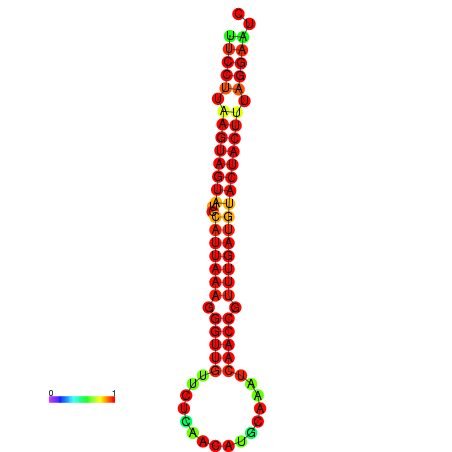
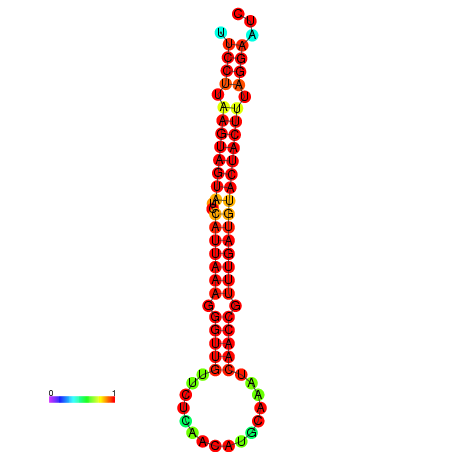
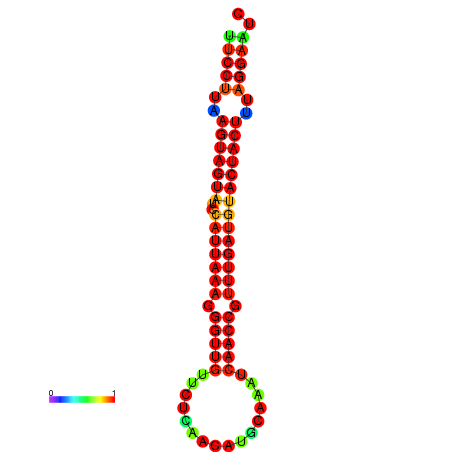
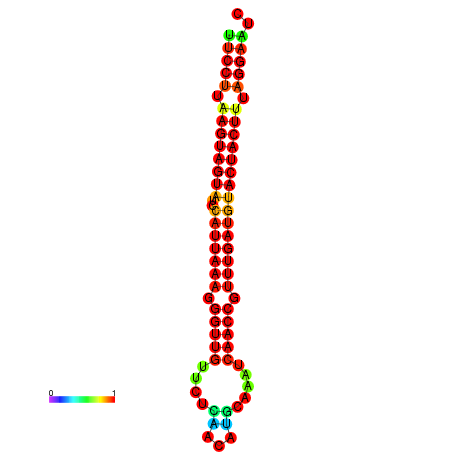
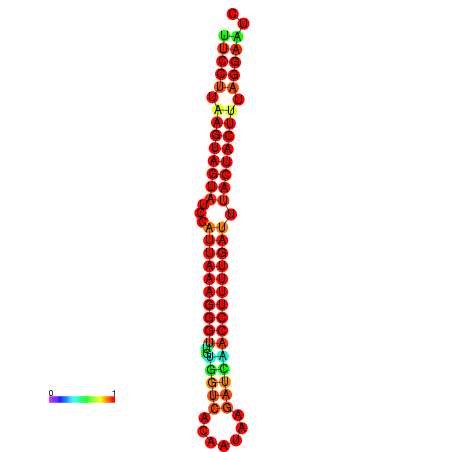
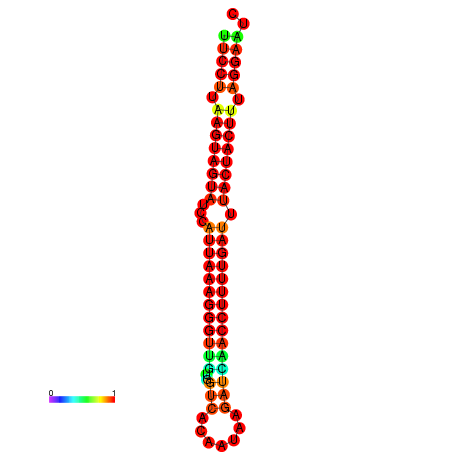
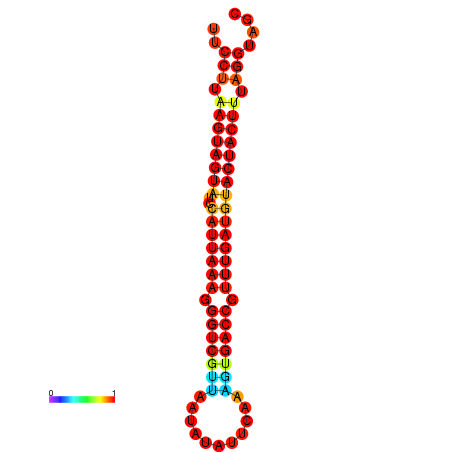
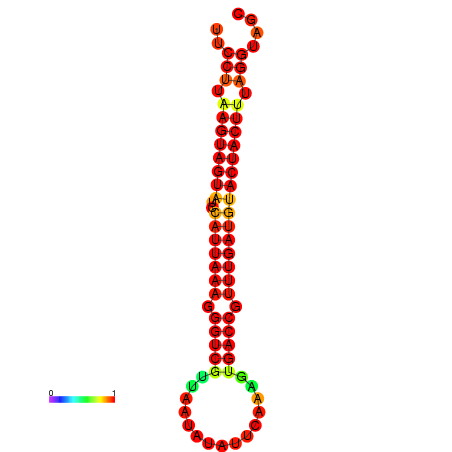
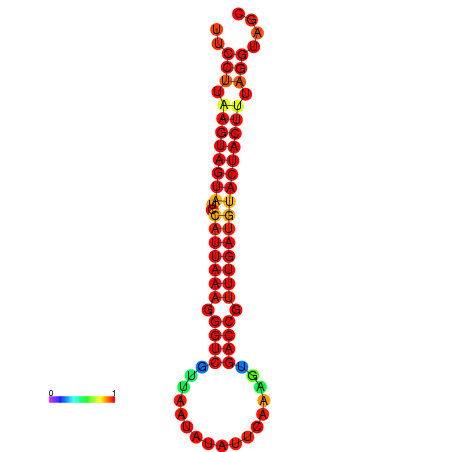
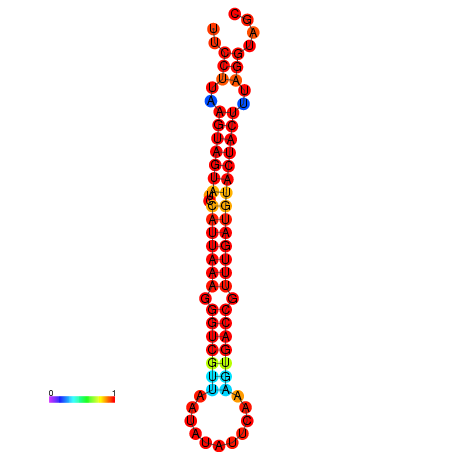
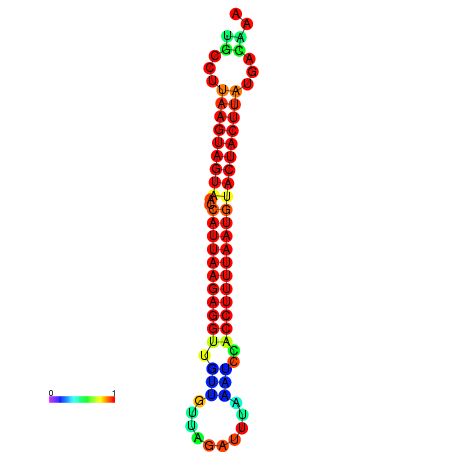
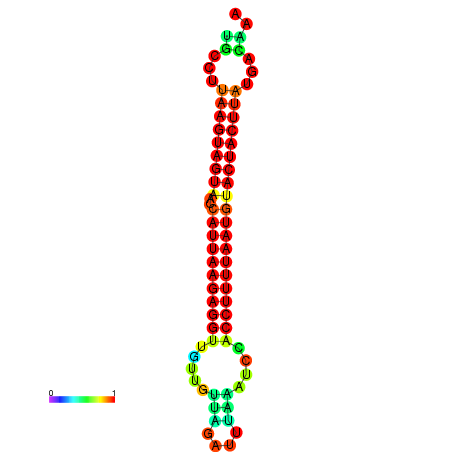
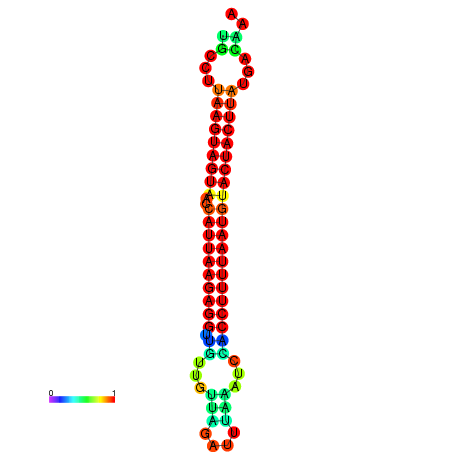
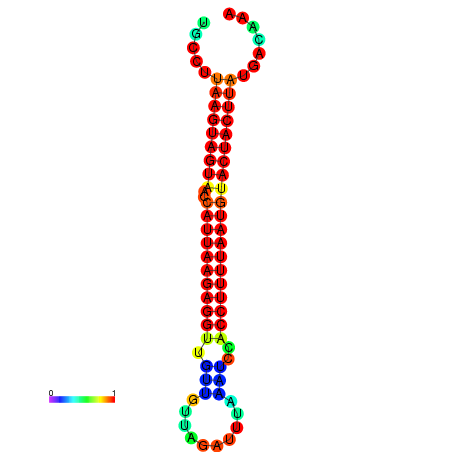
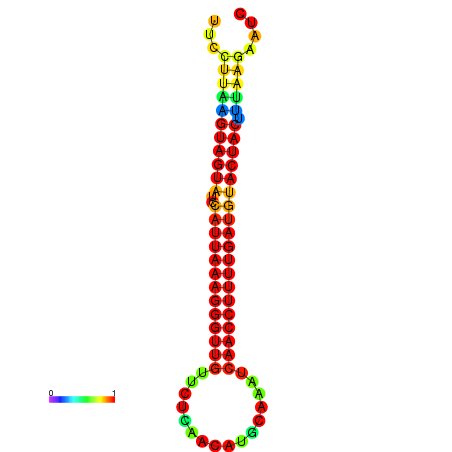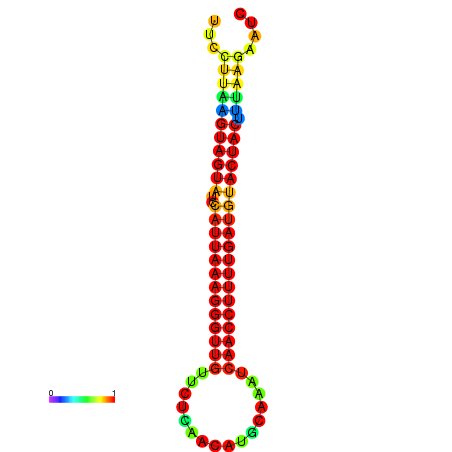| TTGAATAGTGATTTAGTTCTACAATTGCT----GCAGCTTCCTTAAGTAGTATCCATTAAGAGGTCGTCATCTAGCTA-AAATCGACCTTTTGATGTACTACTTTAAAAAAGCACAGTCAAAGTCAACAGAACTG | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ...........................................TAAGTAGTATCCATTAAGAGGTCG.................................................................... | 24 | 1 | 28725.00 | 28725 | 28717 | 8 |
| ..........................................TTAAGTAGTATCCATTAAGAGGTC..................................................................... | 24 | 1 | 559.00 | 559 | 559 | 0 |
| ............................................AAGTAGTATCCATTAAGAGGTCG.................................................................... | 23 | 1 | 551.00 | 551 | 550 | 1 |
| ...........................................TAAGTAGTATCCATTAAGAGGTC..................................................................... | 23 | 1 | 280.00 | 280 | 278 | 2 |
| ............................................AAGTAGTATCCATTAAGAGGTCGT................................................................... | 24 | 1 | 279.00 | 279 | 279 | 0 |
| ...........................................TAAGTAGTATCCATTAAGAGGTCGT................................................................... | 25 | 1 | 96.00 | 96 | 96 | 0 |
| ...........................................TAAGTAGTATCCATTAAGAG........................................................................ | 20 | 1 | 87.00 | 87 | 87 | 0 |
| ...........................................TAAGTAGTATCCATTAAGAGGT...................................................................... | 22 | 1 | 60.00 | 60 | 60 | 0 |
| ...........................................TAAGTAGTATCCATTAAGAGG....................................................................... | 21 | 1 | 51.00 | 51 | 51 | 0 |
| ..........................................TTAAGTAGTATCCATTAAGAGGT...................................................................... | 23 | 1 | 44.00 | 44 | 44 | 0 |
| ...........................................TAAGTAGTATCCATTAAGA......................................................................... | 19 | 1 | 41.00 | 41 | 41 | 0 |
| ..........................................TTAAGTAGTATCCATTAAGAGGTCG.................................................................... | 25 | 1 | 39.00 | 39 | 39 | 0 |
| .............................................AGTAGTATCCATTAAGAGGTCG.................................................................... | 22 | 1 | 23.00 | 23 | 23 | 0 |
| ...............................................TAGTATCCATTAAGAGGTCG.................................................................... | 20 | 1 | 19.00 | 19 | 19 | 0 |
| .....................................................................................ACCTTTTGATGTACTACTTTAA............................ | 22 | 1 | 12.00 | 12 | 12 | 0 |
| ..........................................TTAAGTAGTATCCATTAAGAGG....................................................................... | 22 | 1 | 12.00 | 12 | 12 | 0 |
| ................................................AGTATCCATTAAGAGGTCG.................................................................... | 19 | 1 | 11.00 | 11 | 11 | 0 |
| ..............................................GTAGTATCCATTAAGAGGTCG.................................................................... | 21 | 1 | 10.00 | 10 | 10 | 0 |
| ............................................AAGTAGTATCCATTAAGAGGT...................................................................... | 21 | 1 | 10.00 | 10 | 10 | 0 |
| .....................................................................................ACCTTTTGATGTACTACTTTAAA........................... | 23 | 1 | 9.00 | 9 | 9 | 0 |
| ......................................................................................CCTTTTGATGTACTACTTTAAA........................... | 22 | 1 | 6.00 | 6 | 6 | 0 |
| .....................................................................................ACCTTTTGATGTACTACTTTA............................. | 21 | 1 | 6.00 | 6 | 6 | 0 |
| ..........................................TTAAGTAGTATCCATTAAGA......................................................................... | 20 | 1 | 5.00 | 5 | 5 | 0 |
| ....................................................................................GACCTTTTGATGTACTACTTTA............................. | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ............................................AAGTAGTATCCATTAAGA......................................................................... | 18 | 1 | 4.00 | 4 | 4 | 0 |
| ...........................................TAAGTAGTATCCATTAAG.......................................................................... | 18 | 1 | 3.00 | 3 | 3 | 0 |
| ..........................................TTAAGTAGTATCCATTAAGAG........................................................................ | 21 | 1 | 3.00 | 3 | 3 | 0 |
| ............................................AAGTAGTATCCATTAAGAG........................................................................ | 19 | 1 | 3.00 | 3 | 3 | 0 |
| ............................................AAGTAGTATCCATTAAGAGGTC..................................................................... | 22 | 1 | 3.00 | 3 | 3 | 0 |