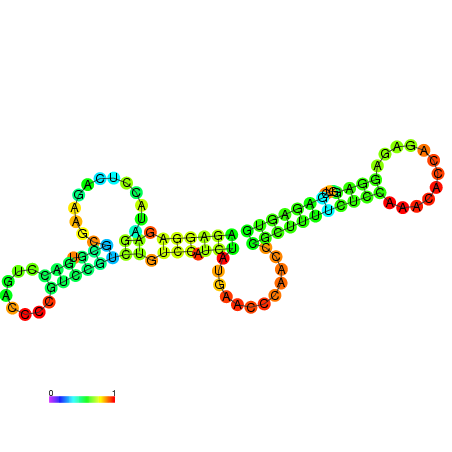
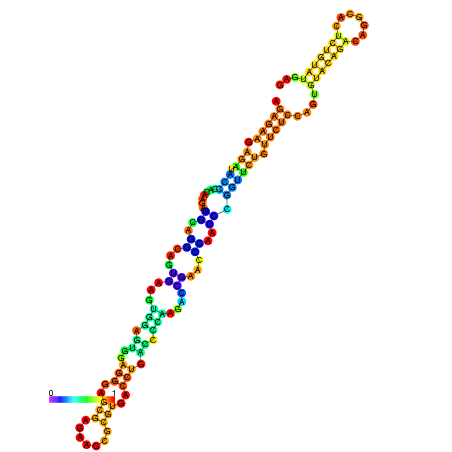
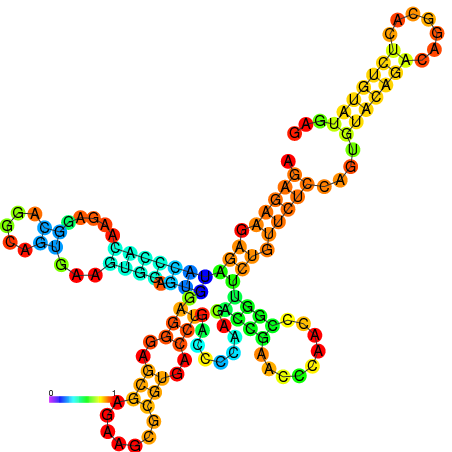
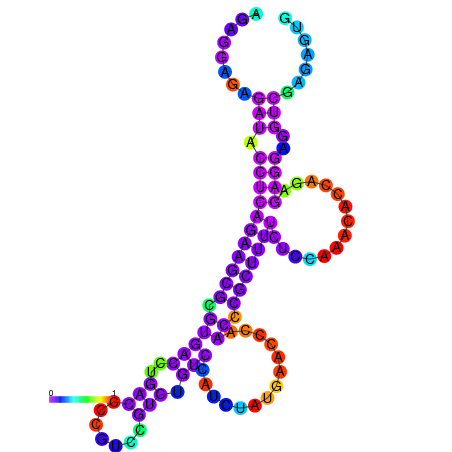| dm3 |
chr2L:12459989-12460115 - |
GAGAACTGCAGGA------GACTTGGAGAGCAGAGATA-----------------------------CCT--------CTGGAGAAGCG--CGTGACCTGACCCCAGCAGGA-----------GAACCCAACCCGCTT----TTCCACCTA----------------G--------------GTGTCTC--T-----------CTCTCTCTTT--ATCTGTTCCTC------AGCTCG |
| droSim1 |
chr2L:12267908-12268042 - |
GAGAACTGCAGGA------GACTTGGAGAGCAGAGATA-----------------------------CCT--------CCGGAGAAGCG--CGTGACCTGACCCCAGCAGGA-----------GAACCCAACCCGCTT----TTCCGCCTA----------------G--------------GTGTCTC--TCTCTCTC---TCTCACTC--TATCTCAGTTCCTC------AGCTCG |
| droSec1 |
super_16:651020-651150 - |
GAGAACTGCAGGA------GACTTGGAGAGCAGAGATA-----------------------------CCT--------CCGGAGAAGCG--CGTGACCTGACCCCAGCAGGA-----------GAACCCAACCCGCTT----TTCCGCCTA----------------G--------------GTGTCTC--T----CTC---TCTCTCTC--TATCTCAGTTCCTC------AGCTCG |
| droYak2 |
chr2L:8897071-8897221 - |
GAGAACTGCAGGAGACGGAGACTTGGAGAGCAGAGAC---------AGAGATGA-----------TACCT--------CTGGAGAAGCG--CGTGACCTGACCCCAGCA--------------GAACCCAACCCGCTT----TTCCGCCTA----------------G--------------ATGTCTC--TATCTCGC---TCTCCCTC--T--CTCTGTTCCTCTTGCACAGCTCG |
| droEre2 |
scaffold_4929:12958952-12959089 + |
GAGAACTGCAGGA------GACTTGGAGAGCAGAGAT---------------GA-----------TACCT--------CTGGAGAAGCG--CGTGACCTGACCCCAGCAGGA-----------GAACCCAGCCCGCTT----TTCCGCCTA----------------G--------------GTGTCTC--TCACTCTC---TCTCTCTC--TCTCTCTTCTCCTC------AGCCCG |
| droAna3 |
scaffold_12916:12931176-12931307 + |
GAGAAATGCATGA------GATTTTGAGAGGAGAGATA-----------------------------CCT--------C---AGAAGCG--CGTGACCTGACCCCGTCCGTCTGTCCATCTATGAACCCAACCCGCTT----TTC---TCCAAACACCAGAGAGGAG-----------------------------GTCGAGAGTG----------------ACGG------TGTGTG |
| dp4 |
chr4_group3:3150562-3150729 - |
GAGAACTGCATGA------GACTTGGAGAGAAGAGATACCCACAA--GAGGCAGGCAGTGAAGTGGAGTGGAGGGAGC---GAGAAGCG--CGTGACCTGACCCCAAGACC------------GAACCCAACCCGGTT----CTGTTCTCCAGTGTACAGACAGGCA-----------------------------CTC---TGTATGAGTA--TGTACCCTCCTT------CGCTC- |
| droPer1 |
super_1:4633245-4633407 - |
GAGAACTGCATGA------GATTTGGAGAGAAGAGATACCCACAA--GAGACGGT---CCG--TGGAGTGGAGGGAGC---GAGAAGCG--CGTGACCTGACCCCAAGACC------------GAACCCAACCCGGTT----CTGTTCTCCAGTGTACAGACAGGCA-----------------------------CTC---TGTATGAGTA--TGTACCCTCCTT------CGCTC- |
| droWil1 |
Unknown |
---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_13246:1657163-1657301 - |
AGAGCGAGCAAGC------AGC------------------------------GTT---TTGTGTCTACCTCAGAAAGC---G--AGACG--CG-----TGAACCCAAATGCG-----------AACCTCGACCCGACTCGAATTCCGACTC----------------AACGCAAACGCTCAACTGTCTG--T------C---TGTCTGTC--T--GTCTGTCTGTC------TGCCTG |
| droMoj3 |
scaffold_6500:4266220-4266364 + |
AAGTAGTAGAAGA------AAAAGAAGAAGCAGCGGC------AACAGAAACGTT---TGGTGTCTACCTCAGAAAGC---G--ACACG--CA----TGAACCCCAAAAACG-----------AACCTCGACCCGACC----GACTACC----------------------------------AATTTCTAT------T---TGTCTGACTCT--GTCTCT-GCTG------TGTGTG |
| droGri2 |
scaffold_15252:15585528-15585616 + |
GCAGA--AGAGTA-AAA--GG---AAGAAGCAGAC----------------------------------------------AAAAGACACGCG--TGTTGAACCCGAAAACGAACCCAACTTCAAATTCAACGCAC---------------------------------------------------------------------------------------TTG------CCTCT- |