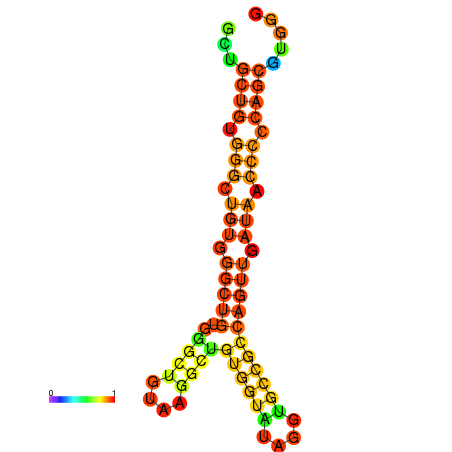
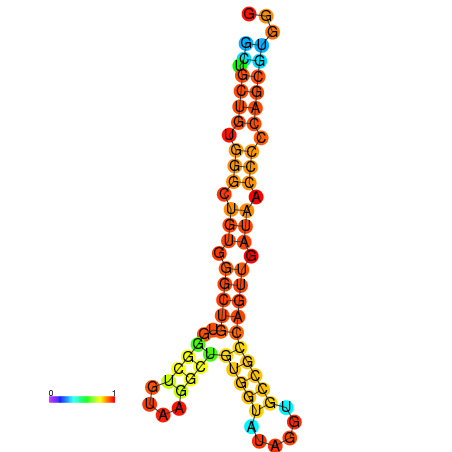
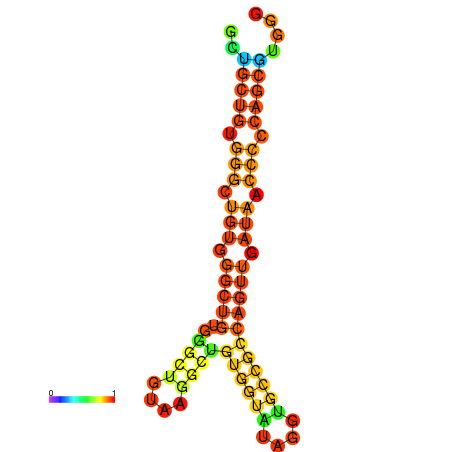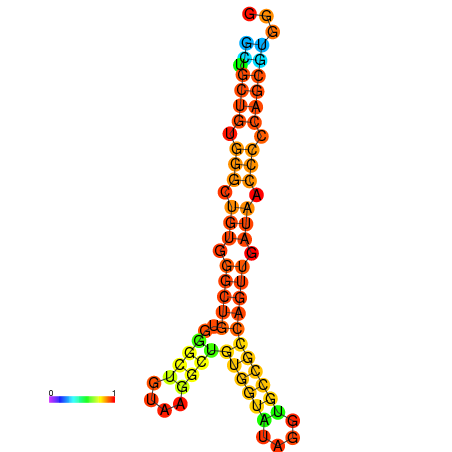| dm3 |
chr2L:6045612-6045728 - |
ATT------T---CGC-TCGGCTGTAACCTGATCCG-----------------CTGCT--GTGGGTTGTGGGCTGTG-------TGGCTGTGG-----TATAGGTGCCGCCAGTTGATAACCCCC-AG-----------------------CGTGGGCA-CGGACCCACTGGTCGTCAACCGC |
| droSim1 |
chr2L:5834370-5834493 - |
ATT------T---CGT-TCGGCTGTAACCTGATCCG-----------------CTGCT--GTGGGCTGTGGGCTGTGGGCTGTAAGGCTGTGG-----TATAGGTGCCGCCAGTTGATAACCCCC-AG-----------------------CGTGGGCA-CGGCCCCACTGGTCGTCAACCGC |
| droSec1 |
super_5:4116543-4116659 - |
ATT------T---CGT-TCGGCTGTAACCTGATCCG-----------------CTGCT--GCGGGTTGTGGGCTGTG-------AGGCTGTGG-----TATAGGTGCCGCCAGTTGATAACCCCC-AG-----------------------CGTGGGCA-CGGCCCCACTGGTCGTCAACCGC |
| droYak2 |
chr2L:13146709-13146824 + |
ATT------C---CGC-TCGGCTGTAACCTGATCCG-----------------CTGCT--GTGGGATGTGGGATGTG--------GGCTGTGG-----TATAGGTGCCGCCAGTTGATAACCCCC-AG-----------------------CGTGGGCA-CGGCCCCACTGGTCGTCAACCGC |
| droEre2 |
scaffold_4929:11075516-11075624 + |
ATT--CC-AT---CAT-TCGGCTGTAACCTGATCCC-----------------CTGCT--GTGG----------------------GCTGTGG-----TATAGGTGCCGCCAGTTGATAACCCCC-AG---------C----ACAG-----CGTGGGCA-CGGC-CCACTGGTCGTCAACCGC |
| droAna3 |
scaffold_12916:8156171-8156273 + |
CAT--CCGATC--TGA-TTGGCAGTAACCTGATCCG-----------------CGGTT--GTG-----------------------GTTGTGG-----TGGAGGTGC---TACTTGATAACCCAC-TG-----------------------CGTGGGCA-CTGGCCTTTCGGTTGTCAACGGC |
| dp4 |
chr4_group1:223208-223319 + |
GGA------T---CGG-GTGGCTGTAACCTGATCTGGGAGCGGTGCAGCAGGGCTGCG--GTGG--------------------------AGT-----TGGAGGTGTCGGCTGTTGATAACCCCC-AG-----------------------CGTAGGCA-CTGGT---ATGGTTGTGAACGGC |
| droPer1 |
super_1:219437-219548 + |
GGA------T---CGG-GTGGCTGTAACCTGATCTGGGAGCGGTGCAGCAGGGCTGCT--GTGG--------------------------AGT-----TGGAGGTGTCGGCTGTTGATAACCCCC-AG-----------------------CGTAGGCA-CTGGT---ATGGTTGTCAACGGC |
| droWil1 |
scaffold_180708:7887454-7887585 + |
GGATTGC-AATTGGTTGTGGGCTGTAACCTGATCTGGA----------------AGCT--ATAG-----------------------ATGTGATGATGCGGAGGTGTCGTCAGATGATAACCCCCCAGGGTAGACAGCTTAGACAT-----CGTAGGCA-TTGGC---CTGGTTGTCAACGGC |
| droVir3 |
scaffold_12963:3941701-3941794 - |
ATT------G---GGCGTCTGCTGTAACTTGAAGCG-----------------CTGATCTG-------------------------------------------------TGGTTGATAACCCCC-TG---------C----CAAGGTGCTCGTAGGCAGCTACCGCTCCGGTTGTCAACGGC |
| droMoj3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
scaffold_15252:1454932-1454939 + |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCAACTAG |