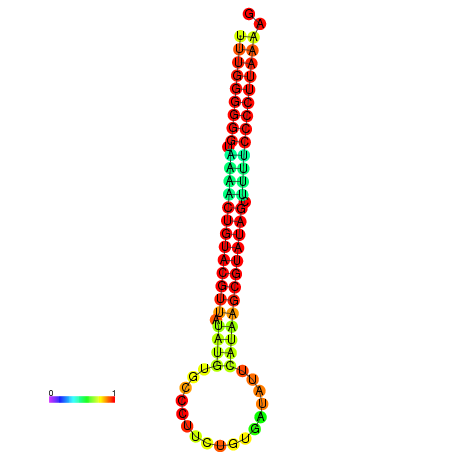
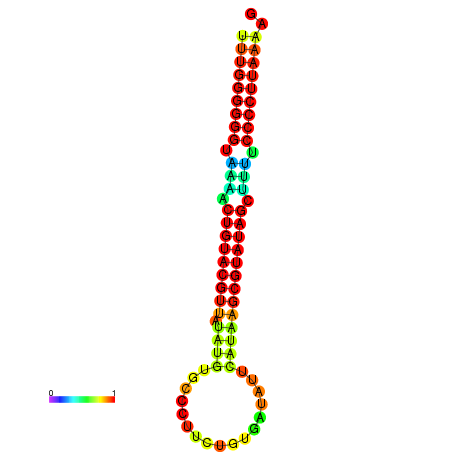
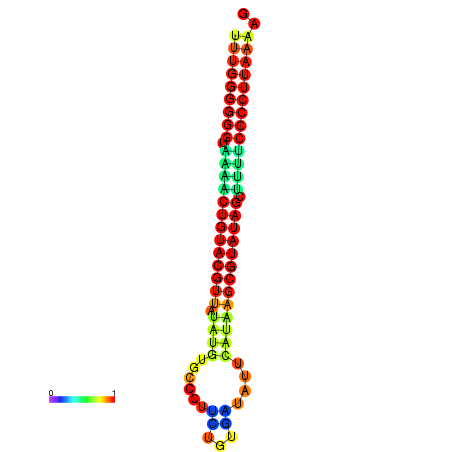
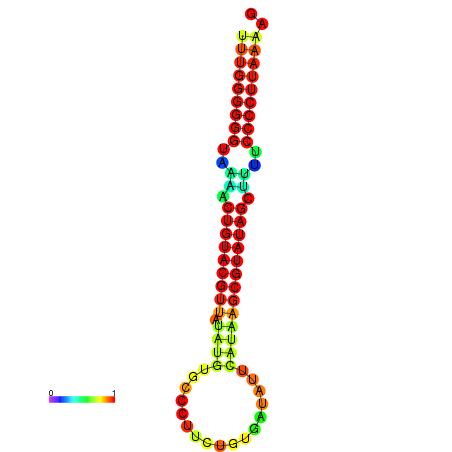
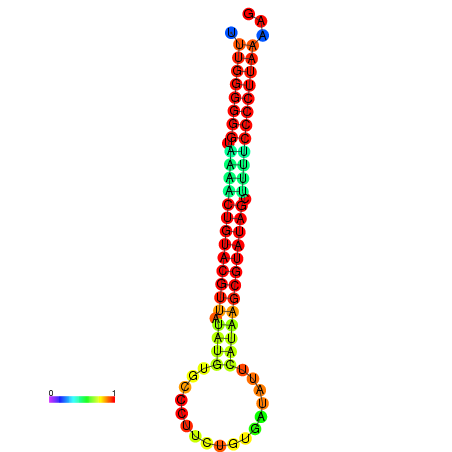
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:243020-243156 - | TAATTGTACAATCTGGA-----CA-----ATATTGCTCAACA-TTTTGGGGGGTAAAACTGTACGTTATATGTGCCCTTCTG-TGATATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTAGAGCTATTGCAACGA----------AAAGT-----GTTTTTC |
| droSim1 | chr2L:248245-248381 - | TACTTGTACAATCTGGA-----CA-----AAATTGCTCAACA-TTTTGGGGGGTGAAACTGTACGTTATATGTGCCCTTCTG-TGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTAGAGCTACTGCAACGA----------AAAGT-----GGTTTTC |
| droSec1 | super_14:240987-241123 - | TACTTGTACAATCTGGA-----CA-----AAATTGCTCAACA-TTTTGGGGGGTGAAACTGTACGTTATATGTGCCCTTCTG-TGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTAGAGCTACTGCAACGA----------AAAGT-----GGTTTTC |
| droYak2 | chr2L:236040-236180 - | TAATTGTACAATCACCATCTGACA-----AAATTGCTCAACA-TTTTAGGGGGTTAAACTGTACGTTATATGTGCCCTTT-T-GTAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGCAACAGCAACAA----------ATAGT-----GTATTTC |
| droEre2 | scaffold_4929:289337-289472 - | TACTTGTAGAACCTG-A-----CA-----AAATTGCTCAACA-TTTTAGGGGGTGAAACTGTACGTTATATGTGTCCTTCTG-TAAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTACAGCAACAGCAACAA----------AAAGT-----GAATCTT |
| droAna3 | scaffold_12943:4147952-4148086 + | TGGGTTTTCTGG--------------------AAGCTCAACCCTTTTGGGGGGTCAAACTGTACGTTCTATGTGGCAATCTT-AGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGCAAAGTCAACAAATCTCAACT-AAAGT-----GTT--TT |
| dp4 | chr4_group1:4362148-4362281 + | TCATTTCCCCAT--------------------CTGCTCAAAA-ATTTGAGGGGTCAAACTGTACGTTCTATGTGACAATATT-CGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAACTTGATGCAACAGCAACAA-TCGCAACAACAATTCTA-------GC |
| droPer1 | super_5:848159-848292 - | TCATTTCCCCAT--------------------CTGCTCAAAA-ATTTGAGGGGTCAAACTGTACGTTCTATGTGACAATATT-CGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAACTTGATGCAACAGCAACAA-TCGCAACAACAATTCTA-------GC |
| droWil1 | scaffold_180764:130106-130233 - | TAT--GTAATAAATTAA-----AGAATCAATTCTACACAACA--TTTATGGGGTTAAACTGTACGTTGTATGTGCAAATCTTAAGAAATCCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGATGTTAAATCAACAA--------------------------- |
| droVir3 | scaffold_12963:8856389-8856513 + | TTAGTGTAGAAAAAGAA-----AAAAACAC--ACACACAAAT-GTTTAGGGGGTCAAACTGTACGTTGTATGTGCAAATG-A-TGAAATTCATAAGCGTATAGCTTTTCCCCTTAAACGTTGATGTGACGACAAC----------------------------- |
| droMoj3 | scaffold_6500:2474475-2474605 - | AATGTGAACAAA--------------------ACACTCAAAT-GTTTAGGGGGTCAAACTGTACGTTGTATGTGTAAACA-A-TGAAATCCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGTGGAATCAACAG----------CAATTTAAGCATAGCTA |
| droGri2 | scaffold_15252:12933990-12934109 - | G--------------GA-----AA-----ATAACACTCAAAT-GTTTAGGGGGTCAAACTGTACGTTATATGTGTAAATG-A-AGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGTGATAGCAACCT----------AAAAA-----GTC--TT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:243020-243156 - | TAATTGTACAATCTGGA-----CA-----ATATTGCTCAACA-TTTTGGGGGGTAAAACTGTACGTTATATGTGCCCTTCTG-TGATATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTAGAGCTATTGCAACGA----------AAAGT-----GTTTTTC |
| droSim1 | chr2L:248245-248381 - | TACTTGTACAATCTGGA-----CA-----AAATTGCTCAACA-TTTTGGGGGGTGAAACTGTACGTTATATGTGCCCTTCTG-TGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTAGAGCTACTGCAACGA----------AAAGT-----GGTTTTC |
| droSec1 | super_14:240987-241123 - | TACTTGTACAATCTGGA-----CA-----AAATTGCTCAACA-TTTTGGGGGGTGAAACTGTACGTTATATGTGCCCTTCTG-TGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTAGAGCTACTGCAACGA----------AAAGT-----GGTTTTC |
| droYak2 | chr2L:236040-236180 - | TAATTGTACAATCACCATCTGACA-----AAATTGCTCAACA-TTTTAGGGGGTTAAACTGTACGTTATATGTGCCCTTT-T-GTAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGCAACAGCAACAA----------ATAGT-----GTATTTC |
| droEre2 | scaffold_4929:289337-289472 - | TACTTGTAGAACCTG-A-----CA-----AAATTGCTCAACA-TTTTAGGGGGTGAAACTGTACGTTATATGTGTCCTTCTG-TAAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTACAGCAACAGCAACAA----------AAAGT-----GAATCTT |
| droAna3 | scaffold_12943:4147952-4148086 + | TGGGTTTTCTGG--------------------AAGCTCAACCCTTTTGGGGGGTCAAACTGTACGTTCTATGTGGCAATCTT-AGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGCAAAGTCAACAAATCTCAACT-AAAGT-----GTT--TT |
| dp4 | chr4_group1:4362148-4362281 + | TCATTTCCCCAT--------------------CTGCTCAAAA-ATTTGAGGGGTCAAACTGTACGTTCTATGTGACAATATT-CGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAACTTGATGCAACAGCAACAA-TCGCAACAACAATTCTA-------GC |
| droPer1 | super_5:848159-848292 - | TCATTTCCCCAT--------------------CTGCTCAAAA-ATTTGAGGGGTCAAACTGTACGTTCTATGTGACAATATT-CGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAACTTGATGCAACAGCAACAA-TCGCAACAACAATTCTA-------GC |
| droWil1 | scaffold_180764:130106-130233 - | TAT--GTAATAAATTAA-----AGAATCAATTCTACACAACA--TTTATGGGGTTAAACTGTACGTTGTATGTGCAAATCTTAAGAAATCCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGATGTTAAATCAACAA--------------------------- |
| droVir3 | scaffold_12963:8856389-8856513 + | TTAGTGTAGAAAAAGAA-----AAAAACAC--ACACACAAAT-GTTTAGGGGGTCAAACTGTACGTTGTATGTGCAAATG-A-TGAAATTCATAAGCGTATAGCTTTTCCCCTTAAACGTTGATGTGACGACAAC----------------------------- |
| droMoj3 | scaffold_6500:2474475-2474605 - | AATGTGAACAAA--------------------ACACTCAAAT-GTTTAGGGGGTCAAACTGTACGTTGTATGTGTAAACA-A-TGAAATCCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGTGGAATCAACAG----------CAATTTAAGCATAGCTA |
| droGri2 | scaffold_15252:12933990-12934109 - | G--------------GA-----AA-----ATAACACTCAAAT-GTTTAGGGGGTCAAACTGTACGTTATATGTGTAAATG-A-AGAAATTCATAAGCGTATAGCTTTTCCCCTTAAAAGTTGAAGTGATAGCAACCT----------AAAAA-----GTC--TT |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-24.8, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
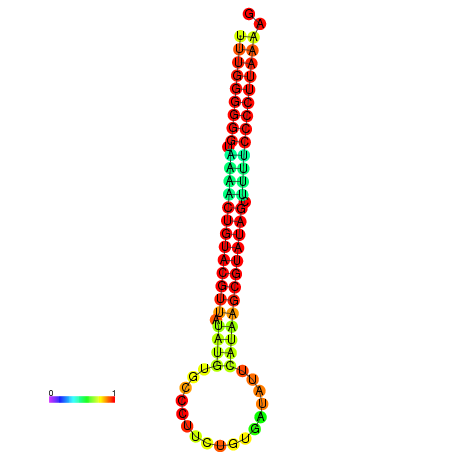 |
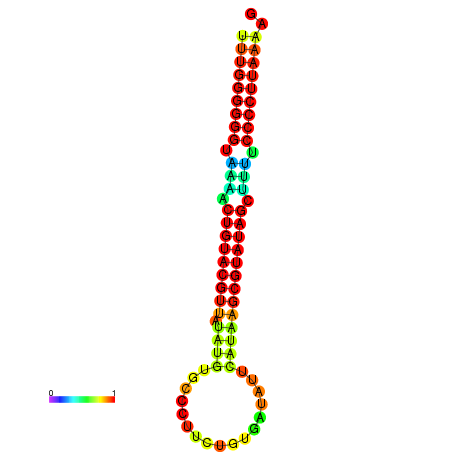 |
 |
 |
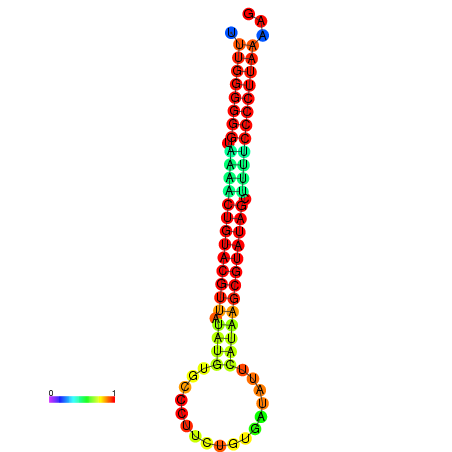 |
| dG=-25.4, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|
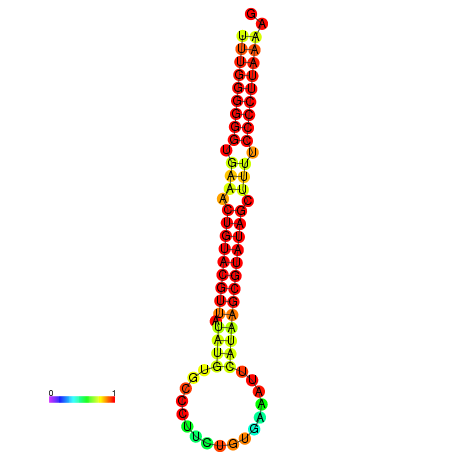 |
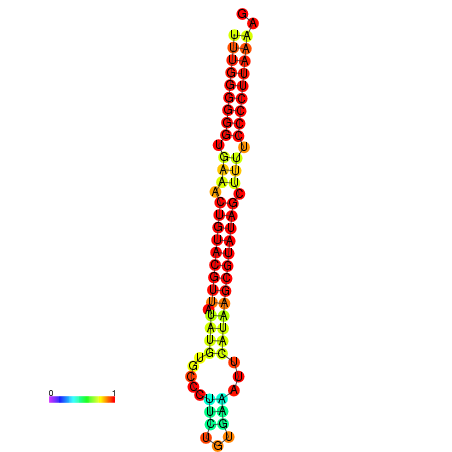 |
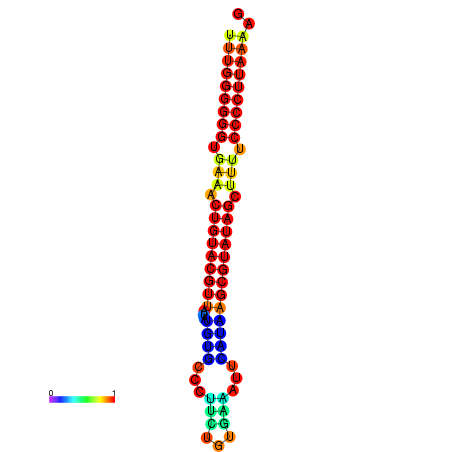 |
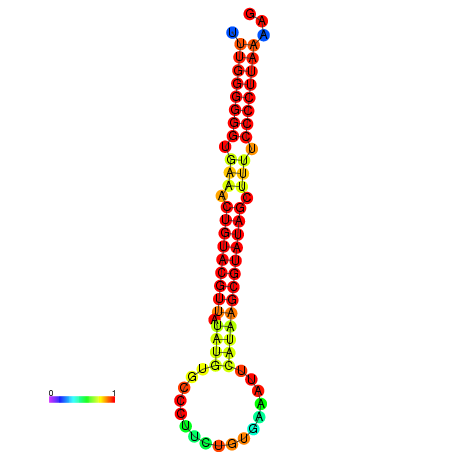 |
| dG=-25.4, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|---|
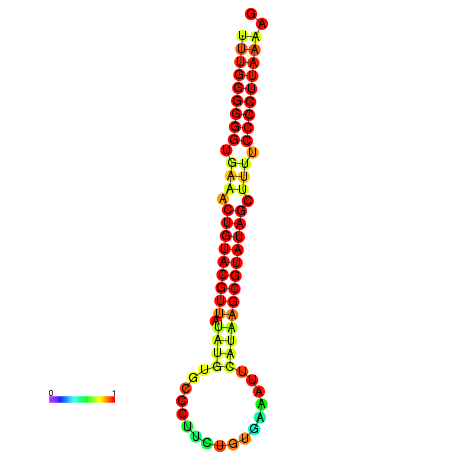 |
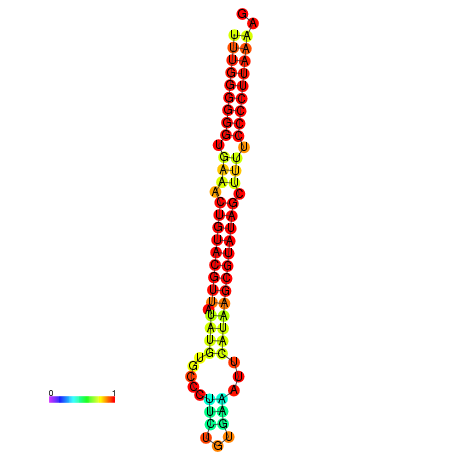 |
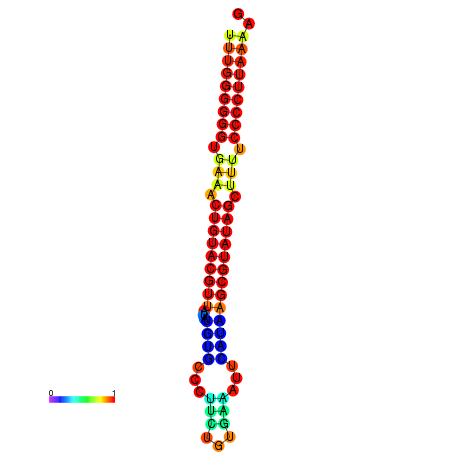 |
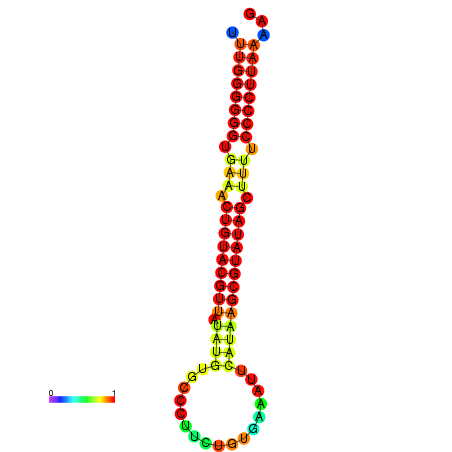 |
| dG=-25.2, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.3, p-value=0.009901 |
|---|---|---|
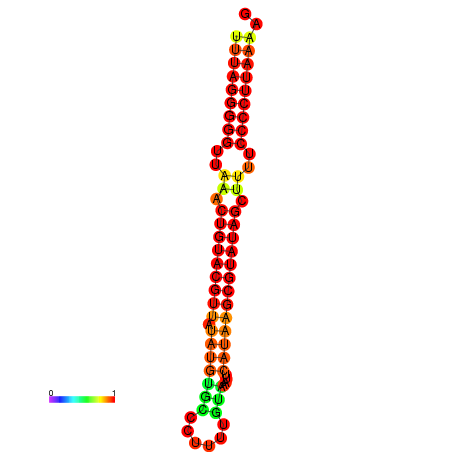 |
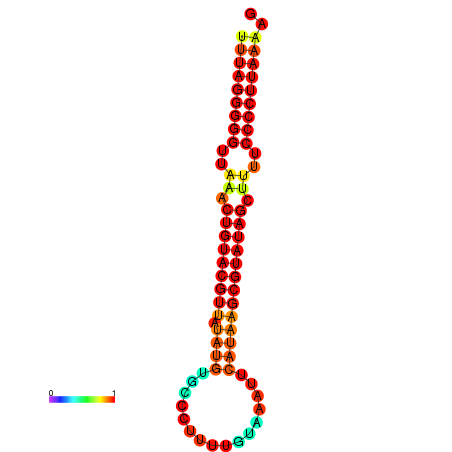 |
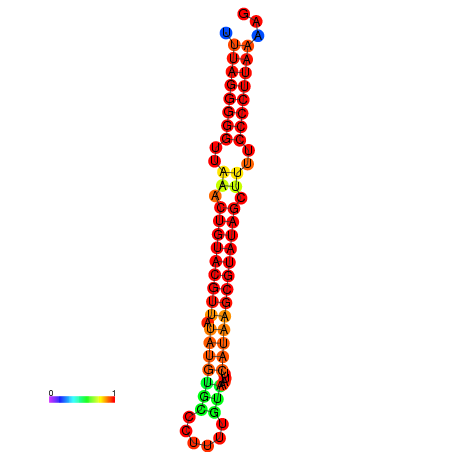 |
| dG=-25.8, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|
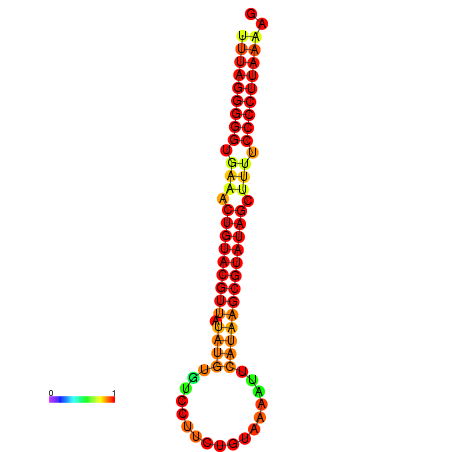 |
 |
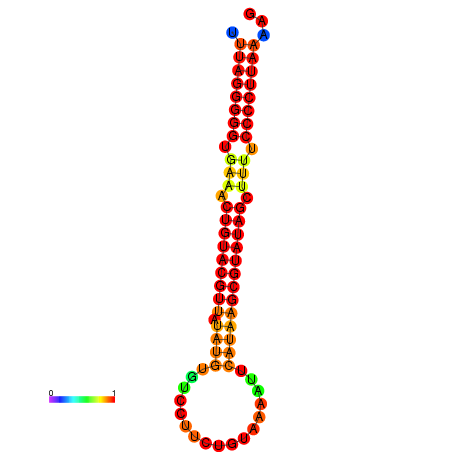 |
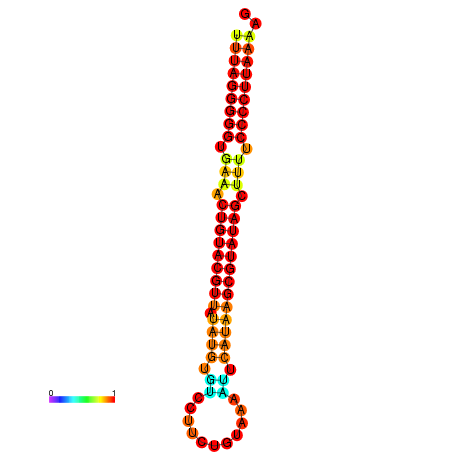
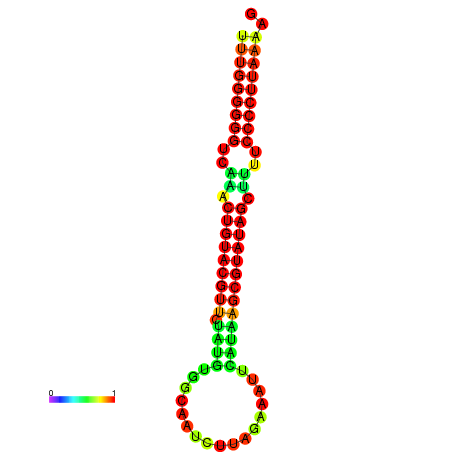
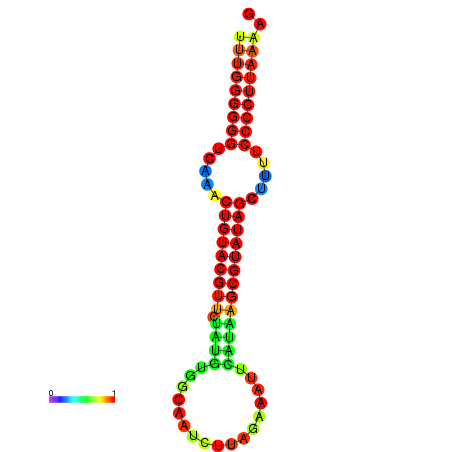
| dG=-23.7, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 | dG=-22.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
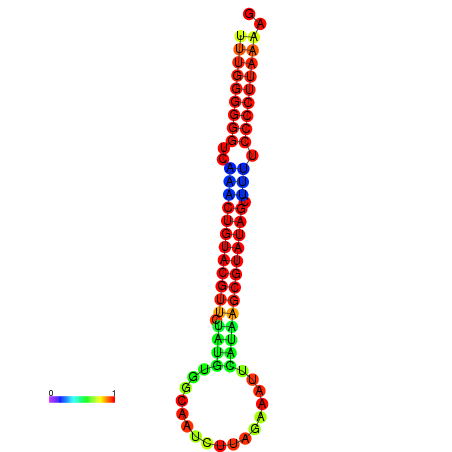 |
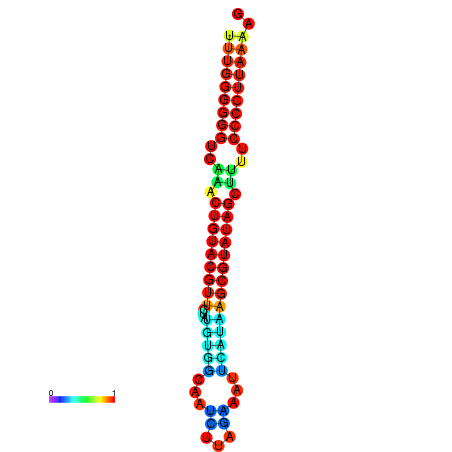 |
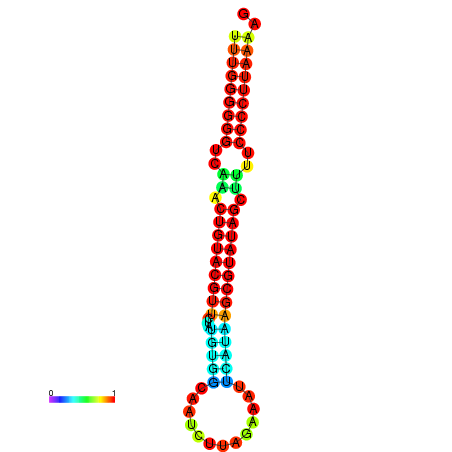 |
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|---|---|---|
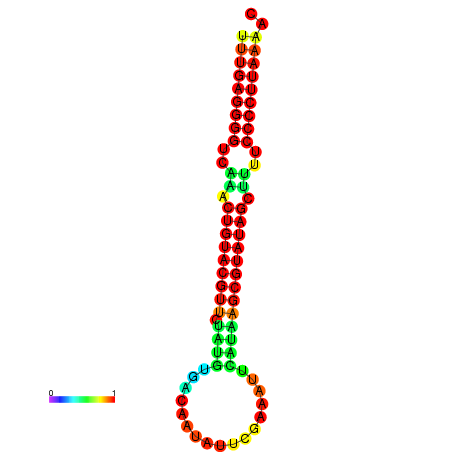 |
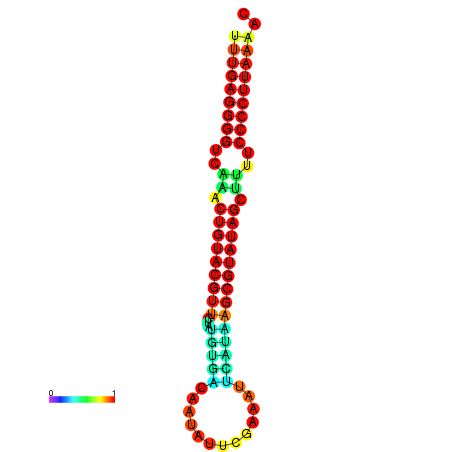 |
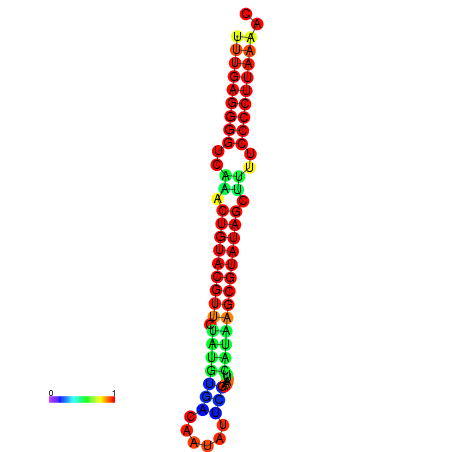 |
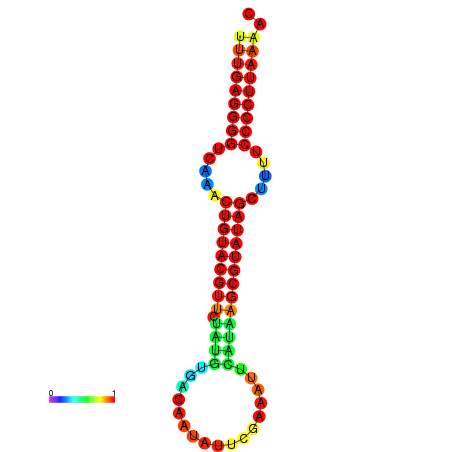 |
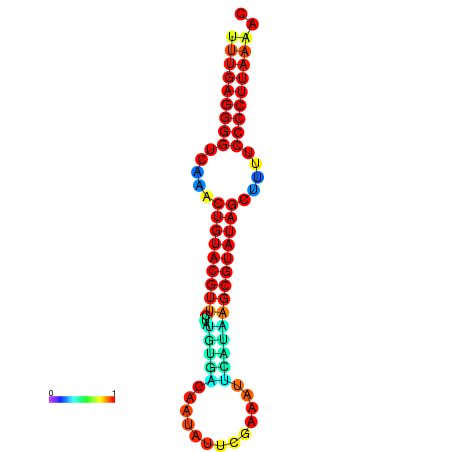 |
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
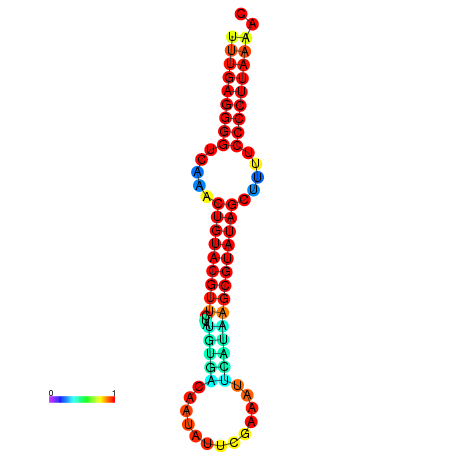 |
| dG=-20.8, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.3, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.8, p-value=0.009901 |
|---|---|---|---|---|
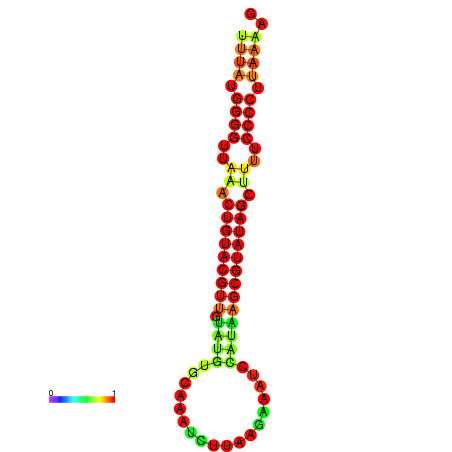 |
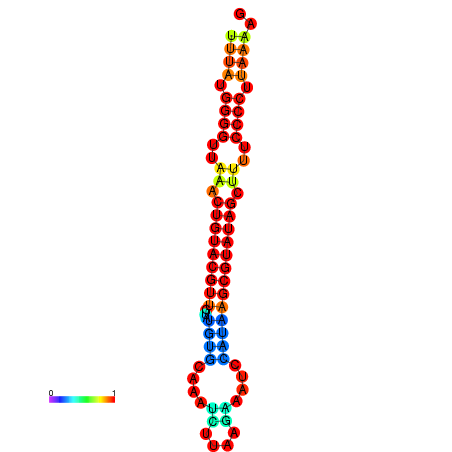 |
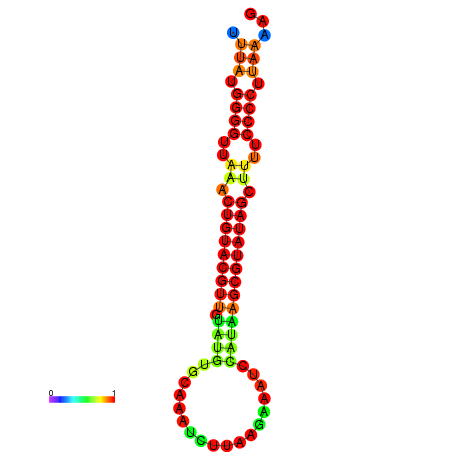
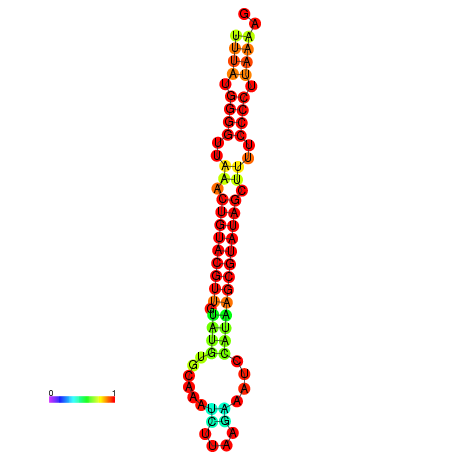 |
 |
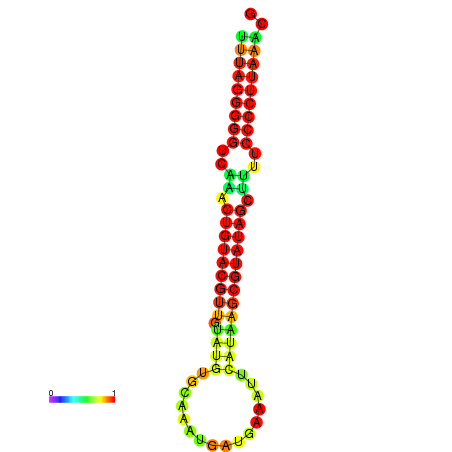
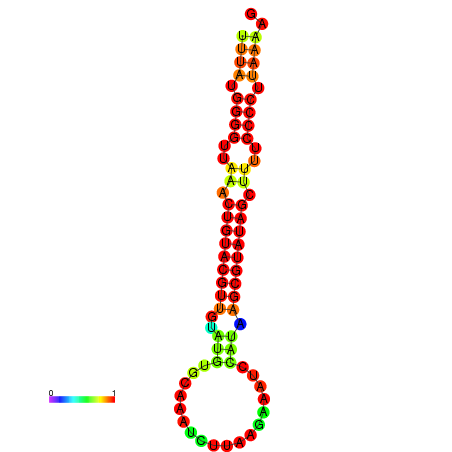 |
| dG=-23.6, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.1, p-value=0.009901 | dG=-23.0, p-value=0.009901 |
|---|---|---|---|---|
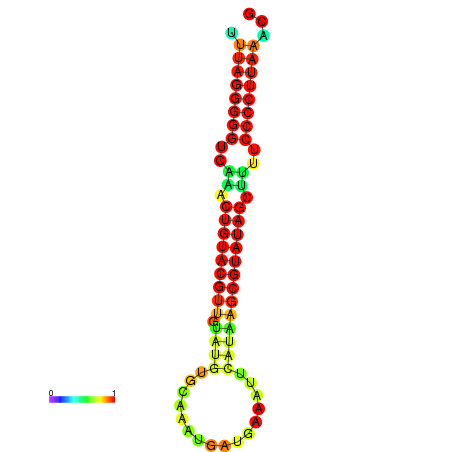
 |
 |
 |
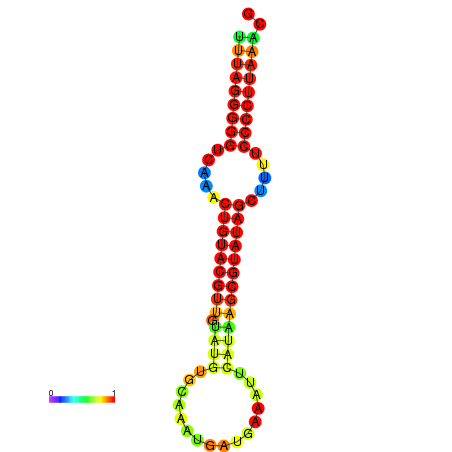 |
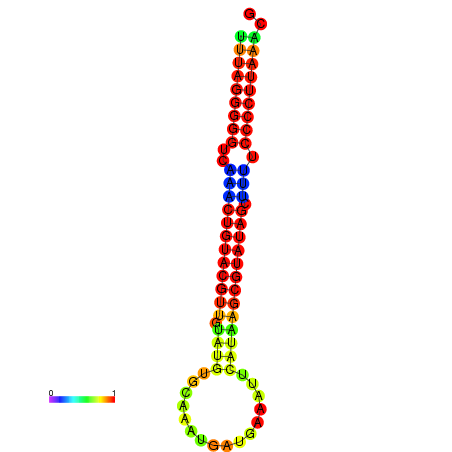 |
| dG=-23.8, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.1, p-value=0.009901 |
|---|---|---|---|---|
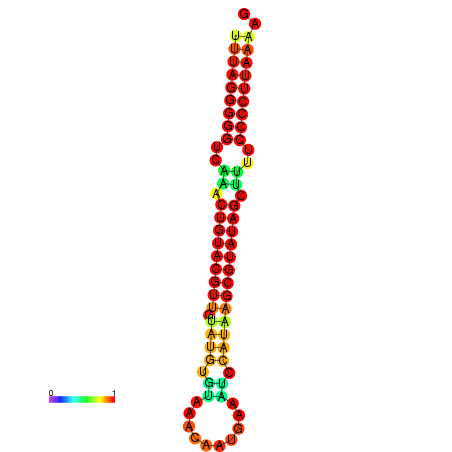 |
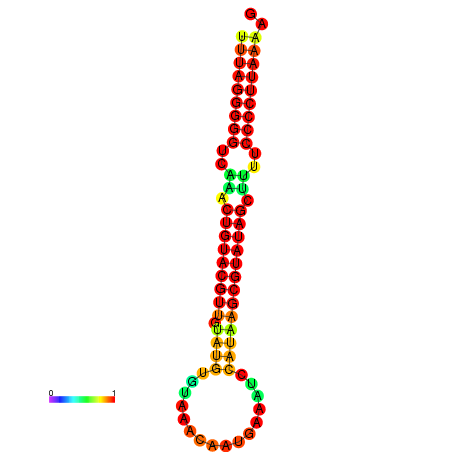 |
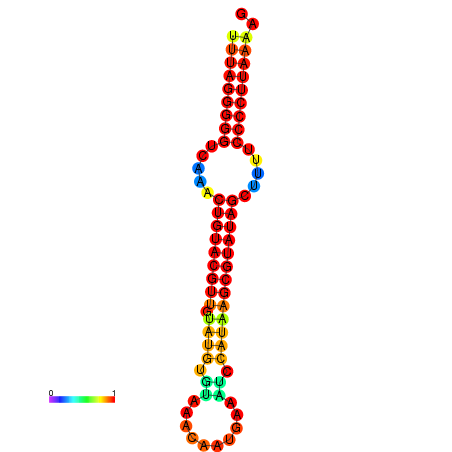 |
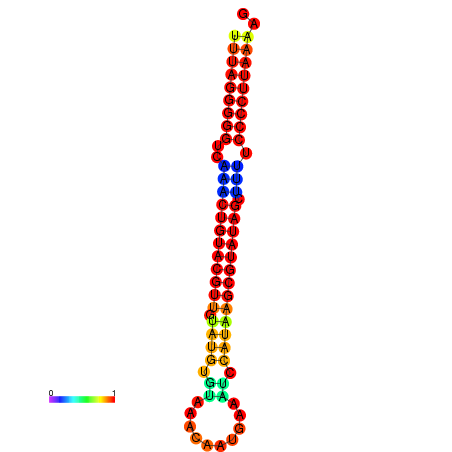 |
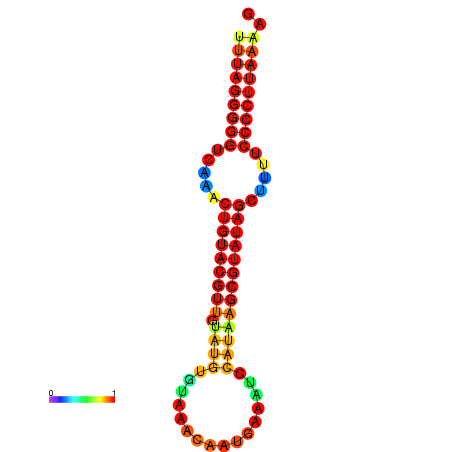 |
| dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.7, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|---|---|---|
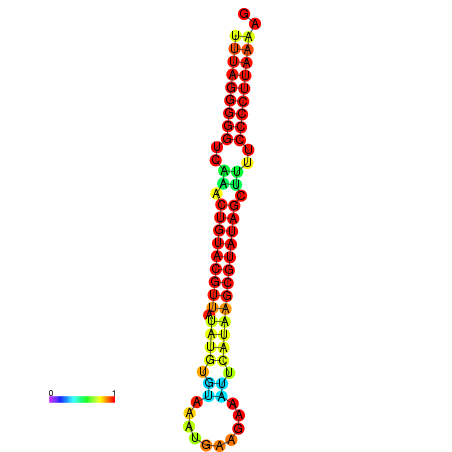 |
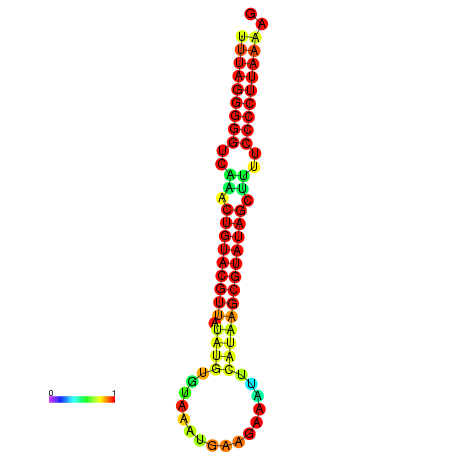 |
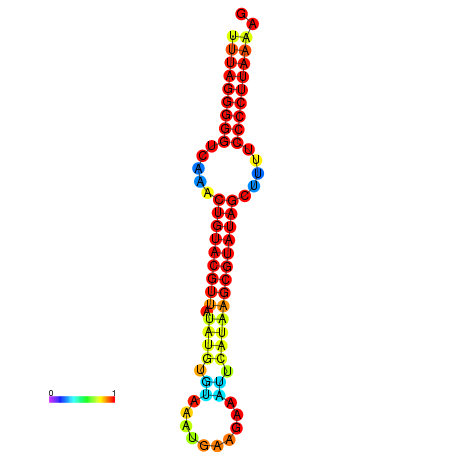 |
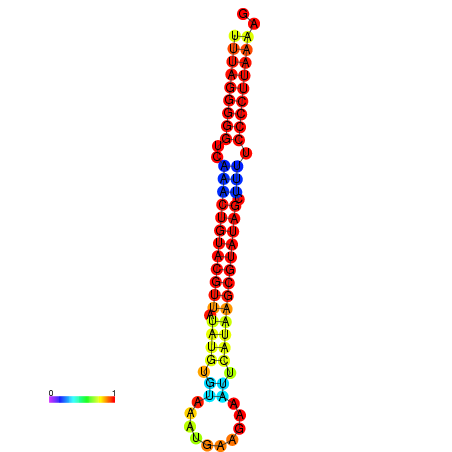 |
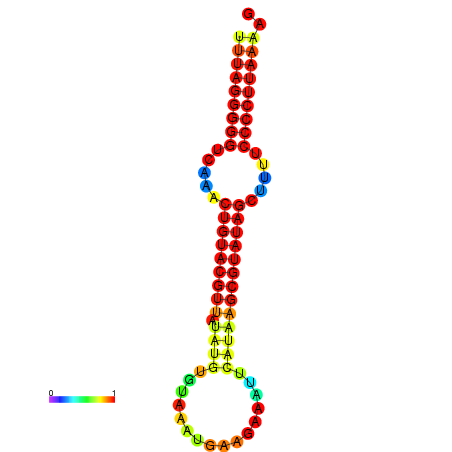 |
Generated: 03/07/2013 at 07:12 PM