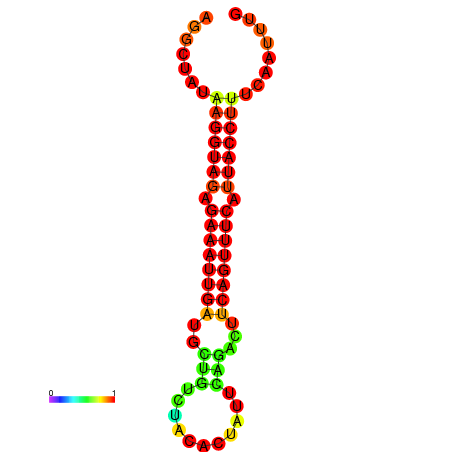
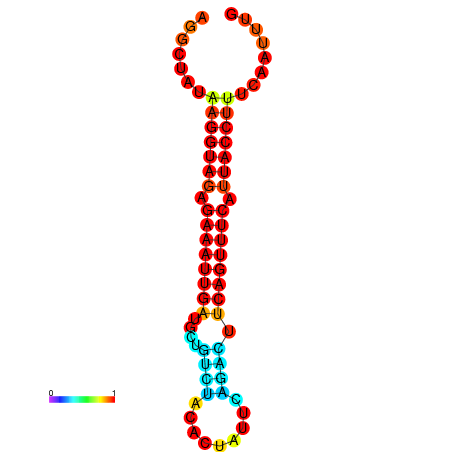
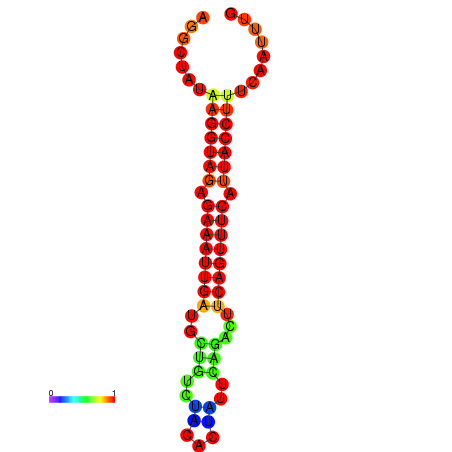
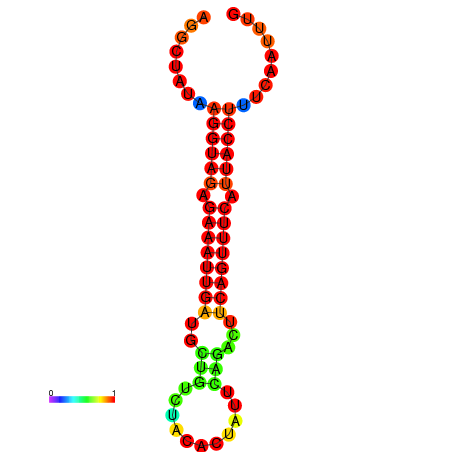
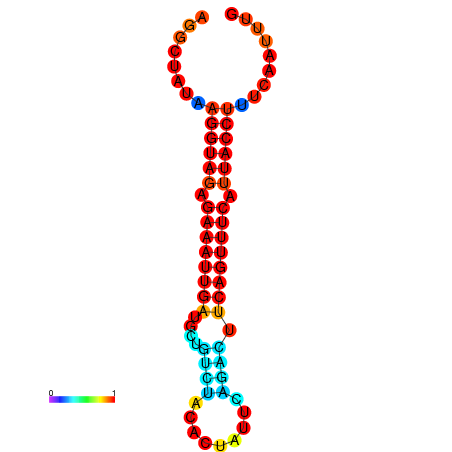
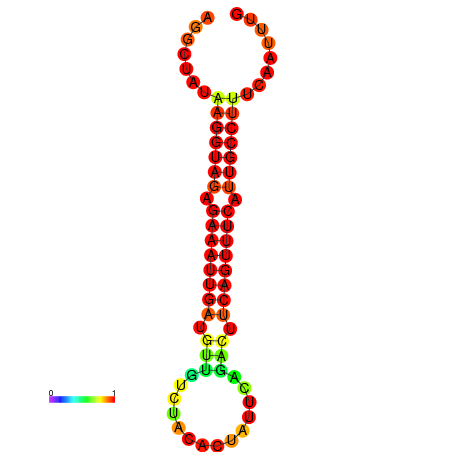
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641283-5641404 + | GAATTCA---ACTACTT---CG-----AT-GGGGC-ACTCAGGCTATAAGGTAGAGAAATTGATGCTGTCTACACTAT--------------T----CAGACTTCAGTTTCATTACCTTTCAATTTGTTTGCCCC-CA--TGCAACAGTGA------------------------------------------TA-----G---------------------------------TA |
| droSim1 | chr2L:5434368-5434488 + | GAATCT-ACAACTACCT---CG-----AT-GGGGC-GCTCAGGCTATAAGGTAGAGAAATTGATGTTGTCTACACTAT--------------T----CAGACTTCAGTTTCATTGCCTTTCAATTTGTTTGCCCC-CC--TGCAACAGTGA------------------------------------------TA----------------------------------------- |
| droSec1 | super_5:3719023-3719143 + | GAATCT-ACAACTACCT---CG-----AT-GGGGC-ATTCAGGCTATAAGGTAGAGAAATTGATGTTGTCTACACTAT--------------T----CAGACTTCAGTTTCATTGCCTTTCAATTTGTTTGCCCC-CA--TGCAACAGTGG------------------------------------------TA----------------------------------------- |
| droYak2 | chr2L:13551914-13552032 - | GAATCCA---ACCAACT---TC-----AT-GGGGC-ACACAAGCTATAAGGTAGAGAAATTGTTGTCTTCCACACTAT--------------T----CAGACATCAGTTTAACTATCTTTCAATTTGTTTGCCCC-CA--CGCAACAGTGA------------------------------------------TA----------------------------------------- |
| droEre2 | scaffold_4929:5729744-5729862 + | GAATCCC---AGTACCT---CC-----TT-GGGGC-ACTCAAGCTATAAGGTAGAAAAATTGTTGTCTTCCACACTAT--------------T----CGGACTGCAGTTTAACTATCTTTCAATTTGTTTGCCCC-CA--CGCAACAGTGA------------------------------------------CA----------------------------------------- |
| droAna3 | scaffold_12916:11085088-11085210 + | GGGCCCA---ACCACCT---GG-----CCGAGGGC-ACTCAAGCTATAAGGTAGAGAAAATAATGTCTTCGTCTCCAA--------------A----TAGACATCAATTTCCCTAGCTTGCATTTTGCCTGCCCC-CA--AGCAACAATGA------------------------------------------TA-----A---------------------------------CA |
| dp4 | chr4_group4:3182204-3182349 - | GAGTGCCACAAATGCCACATGATGCAAAT-GGAGC-AGTCGTGTTTTAAGGTACAGACTTTG---------AGTCTAT--------------TC---TAGGCATCAATTTTCGTATCTTTTAATTCGTCTGGCGC-CAACAGCAACAGCTA--------------------CAAAACGAGTAACGAGGAGAGATA----------------------------------------- |
| droPer1 | super_10:2201135-2201280 - | GAGTGCCACAAATGCCACATGATGCAAAT-GGAGC-AGTCGTGTTTTAAGGTACAGACTTTG---------AGTCTAT--------------TC---TAGGCATCAATTTTCGTATCTTTTAATTCGTCTGCCGC-CAACAGCAACAGCTA--------------------CAAAACGAGTAACGAGGAGAGATA----------------------------------------- |
| droWil1 | scaffold_180708:8625857-8626006 - | GGATAC-ACGTTTTCTATTTAA------TAAGGAC-CCTCAAGTTATAAGGTAGAGAAAATGTTGTCTTTATATT----TGTAACAAT-TGTT----AAGACATCACTTTCACTAGCTTTCAATTTGTCTGCTCCACA--TGCAACAACA-------------------TATAAT-------ACAATCAAACAAA----------------------------------------- |
| droVir3 | scaffold_12963:3615418-3615613 + | ACACAC-ACA--CATAA---GA-----A--AGGGCGCCTCAAATTATAAGGTAGAGAAATTGTTGTCTACT---TTACATATG--------GT----GAGACATCAAT-TCACTAGCTTTCAATTTGTACGCCCA-AA--TGCAACGGAACCATTTTGGAGGAACATCATAT-TT-------AGAATAAGCCACATGATAGGCAGCACATTTAGCATTTGAATGCCGGACTGCTCA |
| droMoj3 | scaffold_6500:14018360-14018470 + | G---------------------------T-AGAGC-GCTCAAGTTATAAGGTAGAGATATTGTTGTCTTTTTGCA----TATAGCACTCTGTTATGAAAGACATCAAT-TCACTAGCTTTCAATTTGTACGCTTA-AA--TGCAGCA----------------------------------------------------------------------------------------- |
| droGri2 | scaffold_15252:3147228-3147335 + | TCATAACACA-----CT---CA--------AGAGC-TCTCAGATTATAAGGTAGAGATTTTGTTGTCTTCTATTATA---------------A----AAGACATCAAT-TCACTAGCTTTCAATTTGTATGCTCA-AA--TGCAACGG---------------------------------------------------------------------------------------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641283-5641404 + | GAATTCA---ACTACTT---CG-----AT-GGGGC-ACTCAGGCTATAAGGTAGAGAAATTGATGCTGTCTACACTAT--------------T----CAGACTTCAGTTTCATTACCTTTCAATTTGTTTGCCCC-CA--TGCAACAGTGA------------------------------------------TA-----G---------------------------------TA |
| droSim1 | chr2L:5434368-5434488 + | GAATCT-ACAACTACCT---CG-----AT-GGGGC-GCTCAGGCTATAAGGTAGAGAAATTGATGTTGTCTACACTAT--------------T----CAGACTTCAGTTTCATTGCCTTTCAATTTGTTTGCCCC-CC--TGCAACAGTGA------------------------------------------TA----------------------------------------- |
| droSec1 | super_5:3719023-3719143 + | GAATCT-ACAACTACCT---CG-----AT-GGGGC-ATTCAGGCTATAAGGTAGAGAAATTGATGTTGTCTACACTAT--------------T----CAGACTTCAGTTTCATTGCCTTTCAATTTGTTTGCCCC-CA--TGCAACAGTGG------------------------------------------TA----------------------------------------- |
| droYak2 | chr2L:13551914-13552032 - | GAATCCA---ACCAACT---TC-----AT-GGGGC-ACACAAGCTATAAGGTAGAGAAATTGTTGTCTTCCACACTAT--------------T----CAGACATCAGTTTAACTATCTTTCAATTTGTTTGCCCC-CA--CGCAACAGTGA------------------------------------------TA----------------------------------------- |
| droEre2 | scaffold_4929:5729744-5729862 + | GAATCCC---AGTACCT---CC-----TT-GGGGC-ACTCAAGCTATAAGGTAGAAAAATTGTTGTCTTCCACACTAT--------------T----CGGACTGCAGTTTAACTATCTTTCAATTTGTTTGCCCC-CA--CGCAACAGTGA------------------------------------------CA----------------------------------------- |
| droAna3 | scaffold_12916:11085088-11085210 + | GGGCCCA---ACCACCT---GG-----CCGAGGGC-ACTCAAGCTATAAGGTAGAGAAAATAATGTCTTCGTCTCCAA--------------A----TAGACATCAATTTCCCTAGCTTGCATTTTGCCTGCCCC-CA--AGCAACAATGA------------------------------------------TA-----A---------------------------------CA |
| dp4 | chr4_group4:3182204-3182349 - | GAGTGCCACAAATGCCACATGATGCAAAT-GGAGC-AGTCGTGTTTTAAGGTACAGACTTTG---------AGTCTAT--------------TC---TAGGCATCAATTTTCGTATCTTTTAATTCGTCTGGCGC-CAACAGCAACAGCTA--------------------CAAAACGAGTAACGAGGAGAGATA----------------------------------------- |
| droPer1 | super_10:2201135-2201280 - | GAGTGCCACAAATGCCACATGATGCAAAT-GGAGC-AGTCGTGTTTTAAGGTACAGACTTTG---------AGTCTAT--------------TC---TAGGCATCAATTTTCGTATCTTTTAATTCGTCTGCCGC-CAACAGCAACAGCTA--------------------CAAAACGAGTAACGAGGAGAGATA----------------------------------------- |
| droWil1 | scaffold_180708:8625857-8626006 - | GGATAC-ACGTTTTCTATTTAA------TAAGGAC-CCTCAAGTTATAAGGTAGAGAAAATGTTGTCTTTATATT----TGTAACAAT-TGTT----AAGACATCACTTTCACTAGCTTTCAATTTGTCTGCTCCACA--TGCAACAACA-------------------TATAAT-------ACAATCAAACAAA----------------------------------------- |
| droVir3 | scaffold_12963:3615418-3615613 + | ACACAC-ACA--CATAA---GA-----A--AGGGCGCCTCAAATTATAAGGTAGAGAAATTGTTGTCTACT---TTACATATG--------GT----GAGACATCAAT-TCACTAGCTTTCAATTTGTACGCCCA-AA--TGCAACGGAACCATTTTGGAGGAACATCATAT-TT-------AGAATAAGCCACATGATAGGCAGCACATTTAGCATTTGAATGCCGGACTGCTCA |
| droMoj3 | scaffold_6500:14018360-14018470 + | G---------------------------T-AGAGC-GCTCAAGTTATAAGGTAGAGATATTGTTGTCTTTTTGCA----TATAGCACTCTGTTATGAAAGACATCAAT-TCACTAGCTTTCAATTTGTACGCTTA-AA--TGCAGCA----------------------------------------------------------------------------------------- |
| droGri2 | scaffold_15252:3147228-3147335 + | TCATAACACA-----CT---CA--------AGAGC-TCTCAGATTATAAGGTAGAGATTTTGTTGTCTTCTATTATA---------------A----AAGACATCAAT-TCACTAGCTTTCAATTTGTATGCTCA-AA--TGCAACGG---------------------------------------------------------------------------------------- |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-17.0, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.5, p-value=0.009901 | dG=-16.3, p-value=0.009901 | dG=-16.2, p-value=0.009901 |
|---|---|---|---|---|
 |
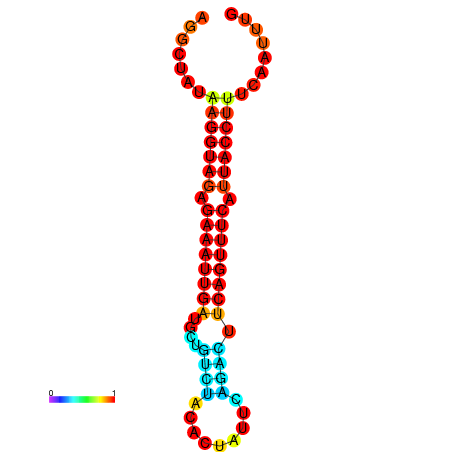 |
 |
 |
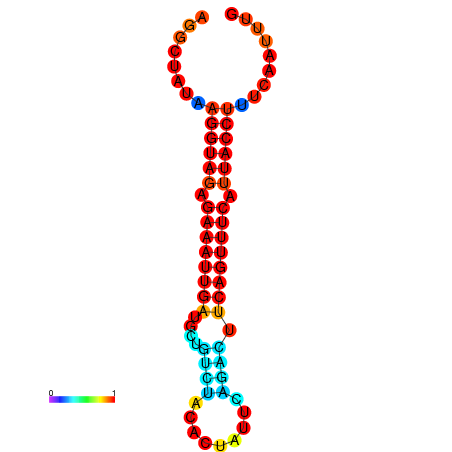 |
| dG=-17.5, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.8, p-value=0.009901 |
|---|---|---|
 |
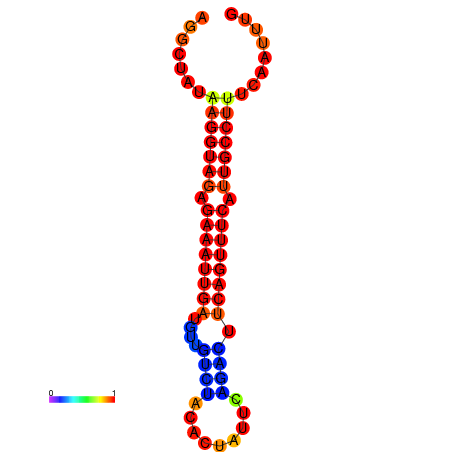 |
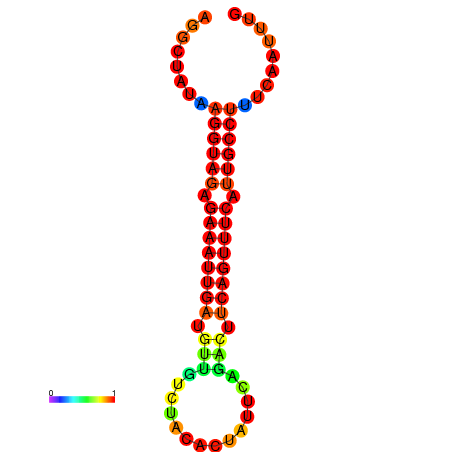 |
| dG=-17.5, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.8, p-value=0.009901 |
|---|---|---|
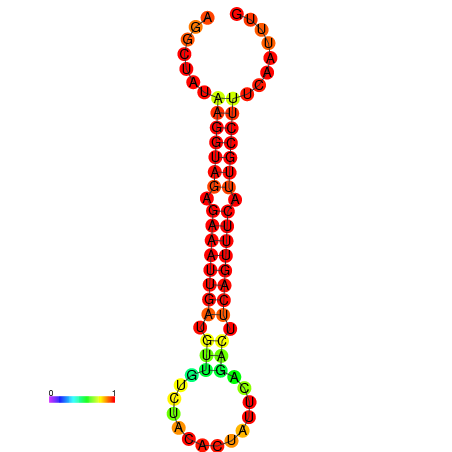 |
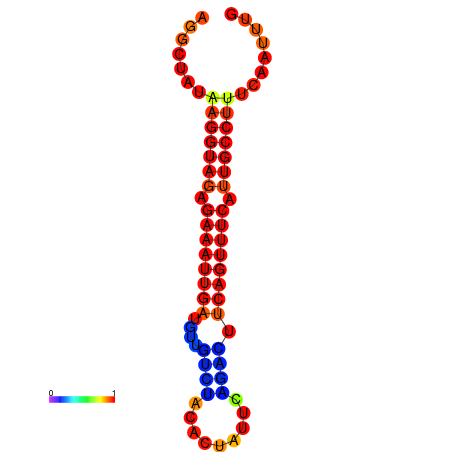 |
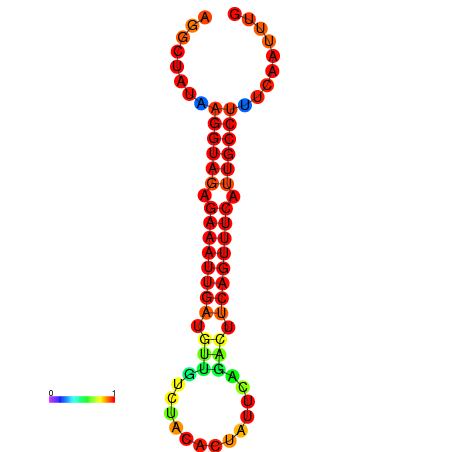 |
| dG=-15.7, p-value=0.009901 | dG=-15.0, p-value=0.009901 |
|---|---|
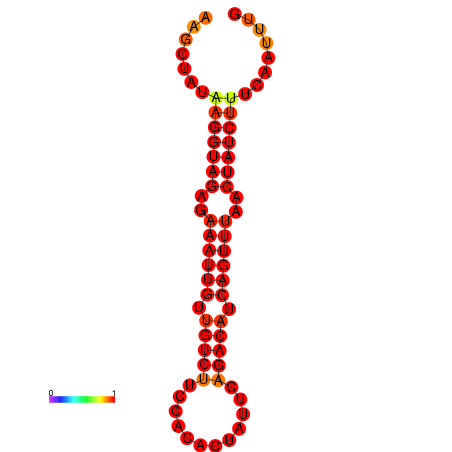 |
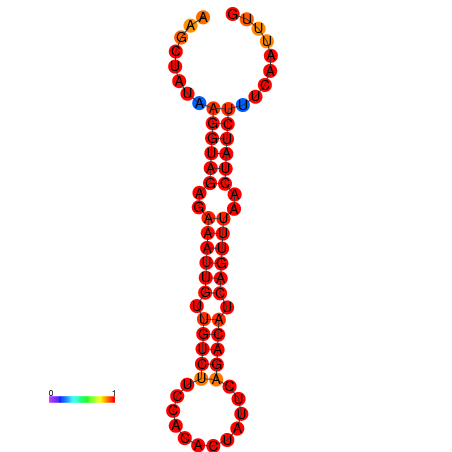 |
| dG=-14.9, p-value=0.009901 | dG=-14.2, p-value=0.009901 |
|---|---|
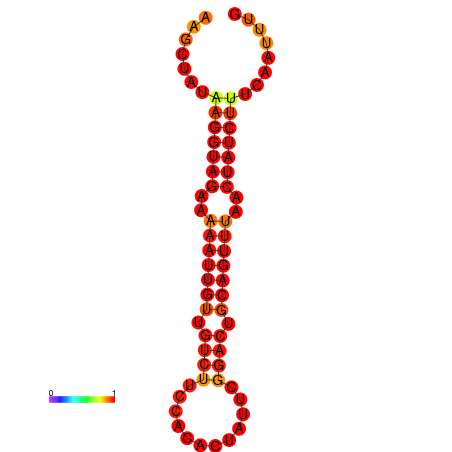 |
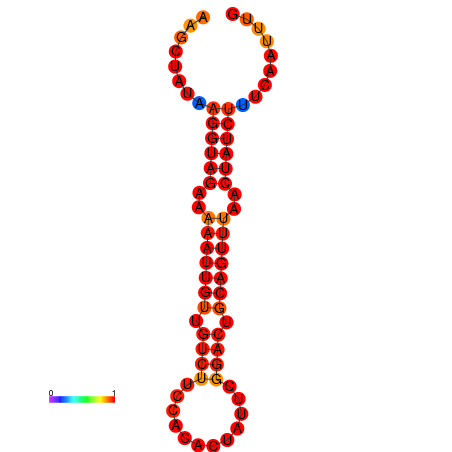 |
| dG=-13.7, p-value=0.009901 | dG=-13.2, p-value=0.009901 |
|---|---|
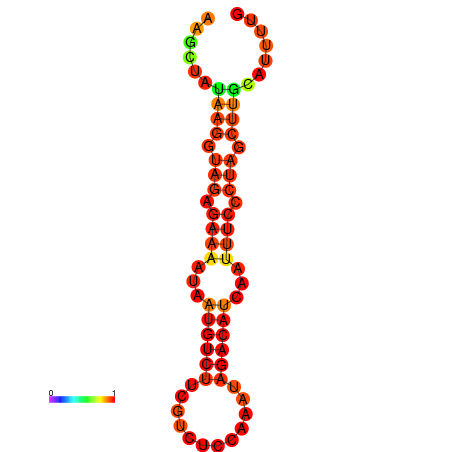 |
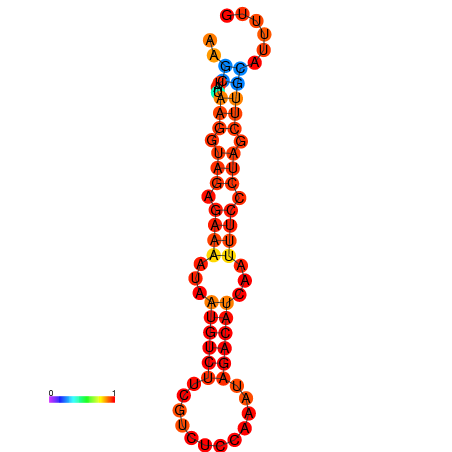 |
| dG=-14.4, p-value=0.009901 | dG=-13.6, p-value=0.009901 | dG=-13.4, p-value=0.009901 |
|---|---|---|
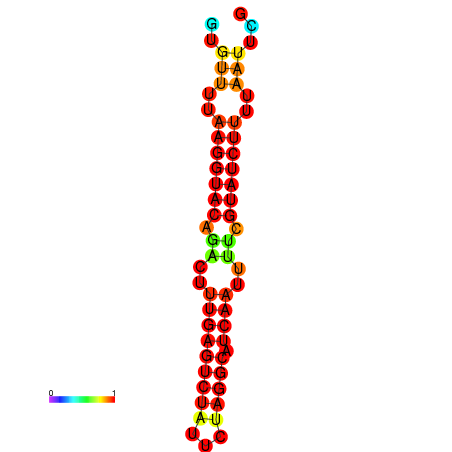 |
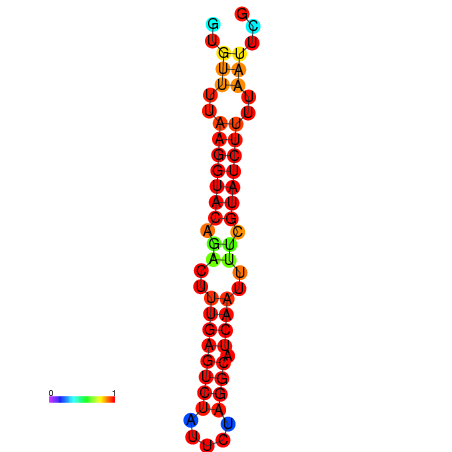 |
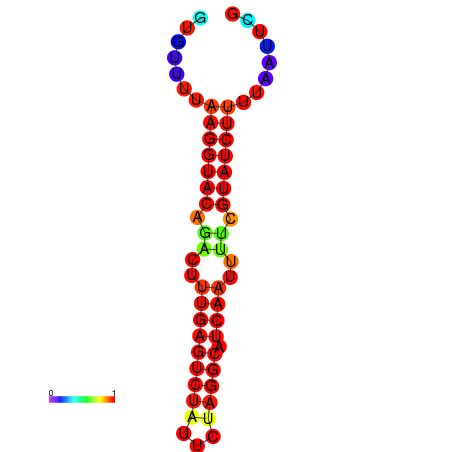 |
| dG=-14.4, p-value=0.009901 | dG=-13.6, p-value=0.009901 | dG=-13.4, p-value=0.009901 |
|---|---|---|
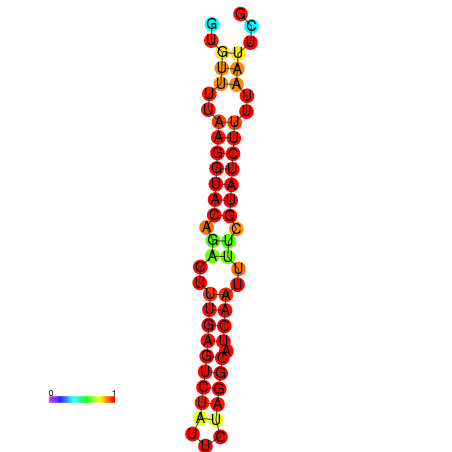 |
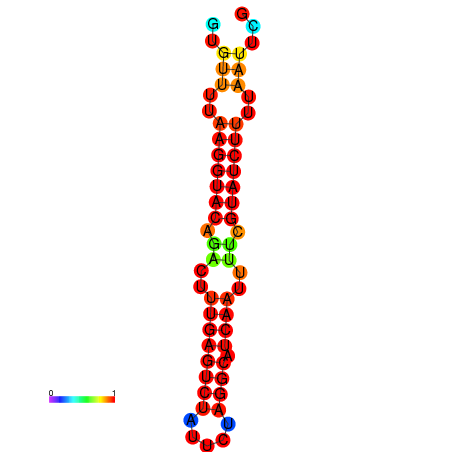 |
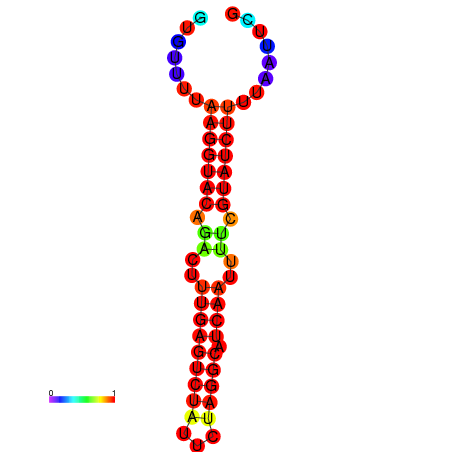 |
| dG=-19.5, p-value=0.009901 | dG=-18.8, p-value=0.009901 | dG=-18.8, p-value=0.009901 |
|---|---|---|
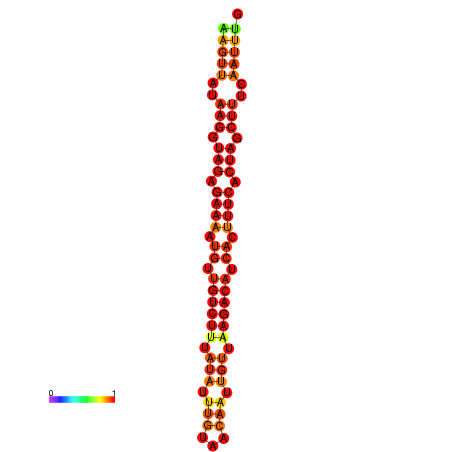 |
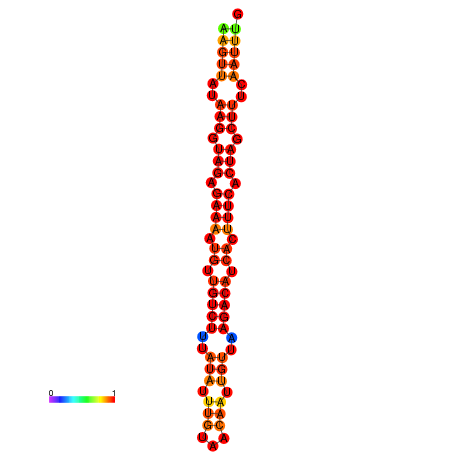 |
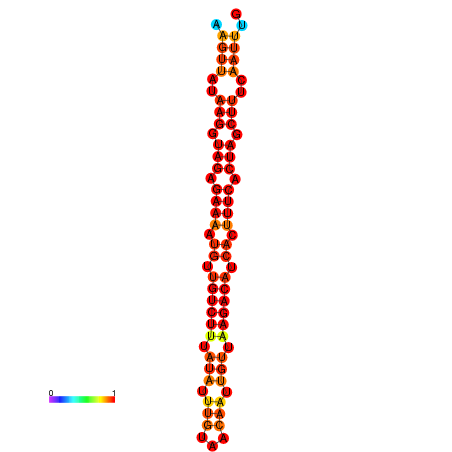 |
| dG=-17.9, p-value=0.009901 | dG=-17.9, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.2, p-value=0.009901 |
|---|---|---|---|---|
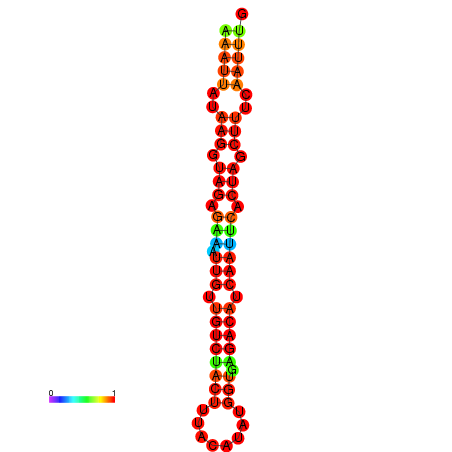 |
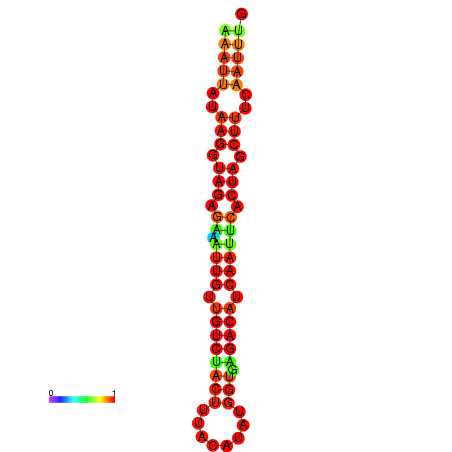 |
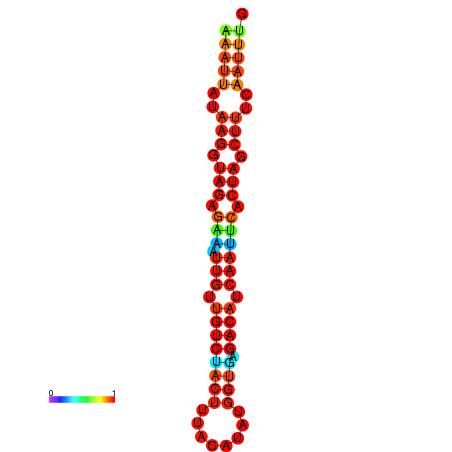 |
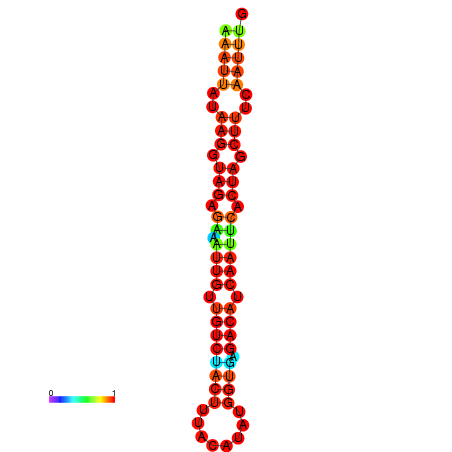 |
 |
| dG=-20.5, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-19.8, p-value=0.009901 | dG=-19.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
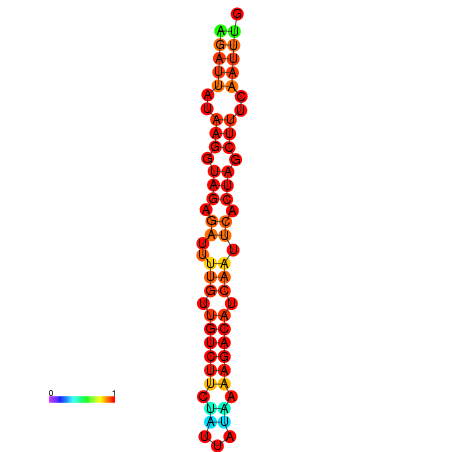
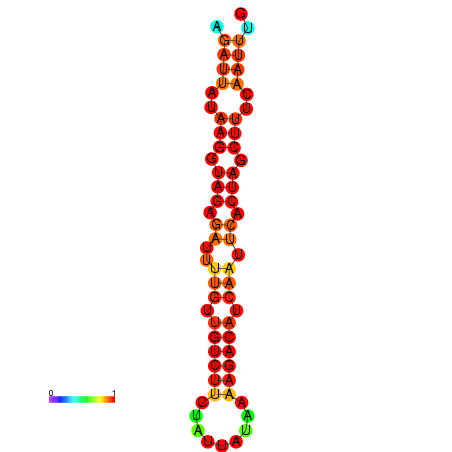
| dG=-14.3, p-value=0.009901 | dG=-14.0, p-value=0.009901 | dG=-13.9, p-value=0.009901 | dG=-13.6, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
Generated: 03/07/2013 at 07:10 PM