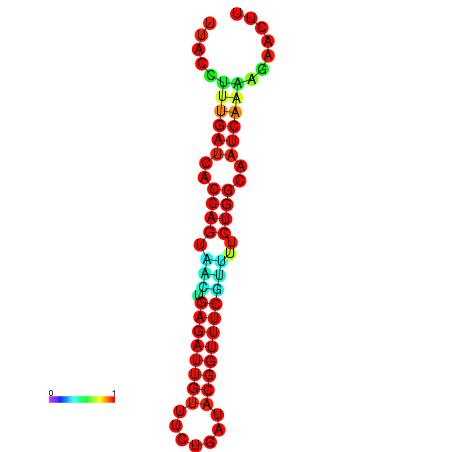
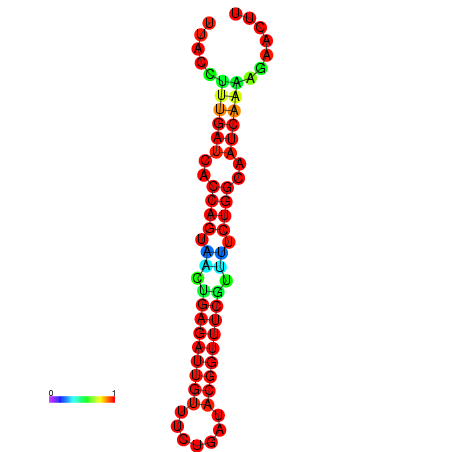

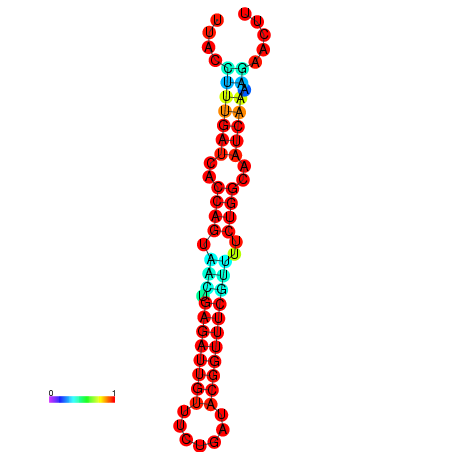

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641176-5641294 + | GTAGTGAGCTC--GT-------T------ATCAAG-----GGCCGAGTTACCTTTGATCACCAGTAACTGAGATT-GT--TTCTGATA---CGGTTTCGTTTTCTGGCAATCAA-AAGAACTTGGACTCGA-CTTGAA----------------------------------------------------------TTCAACTAC |
| droSim1 | chr2L:5434262-5434381 + | GTAATGTGCTC--GT-------T------ATCATG-----GGCCGAGTTACCTTTGATCATCAGTAAGTGAGATT-GT--TTCTGATA---CGATTTCGTTTTCTGGCAATCAA-AGGAACTCGA-CTCGA-CTTGAA-TC-------------------------------------------------------TACAACTAC |
| droSec1 | super_5:3718917-3719036 + | GTAATGTACTC--TT-------T------ATCAAG-----GCCCGAGTTACCTTTGATCACCAGTAAGTGAGATT-GT--TTCTGATA---CGATTTCGTTTTCTGGCAATCAA-AGGAACTCGA-CTCGA-CTTGAA-TC-------------------------------------------------------TACAACTAC |
| droYak2 | chr2L:13552021-13552135 - | GAAAA----TCTAAT---------------TCATG-----GGTCGAGTTACCATTGATCACCGGTTACTGAGGTT-GT--TCCTTCTT---TGGCTTCGTTTTCTGGCGATCAA-AAGAACTCGA-CTCGA-CTCGAA----------------------------------------------------------TCCAACCAA |
| droEre2 | scaffold_4929:5729649-5729755 + | TG-----------AT---------------TCATG-----GGTCGAGTTACCATTGATCACCGGTTACTGAGGTT-GT--GCCACATT---CGGCTTCGTTTCCTGGCGATCAA-AAGAACTCGA-CTCGG-CTCGAA----------------------------------------------------------TCCCAGTAC |
| droAna3 | scaffold_12916:11084979-11085099 + | ACAAAGGACTA--CAGGAAAGCG------ACT-TA----AGTCCGAGCACTATTTGATCACTGGTTACTGAGGTT-GT--TTTGTCAT---AAGCTTCGTTTTCTGGCGATCAA-ATACGCTCGA-CTT-----TGGG----------------------------------------------------------CCCAACCAC |
| dp4 | chr4_group4:3182338-3182458 - | GAGGAGAGCTG--TT---------------TCGCCGGACAGTCAGAGTTTGATTTGATCTCTAGTTACTGGGCTTTGA--TTCGAAATAAGTAGCTTATTATTCTAGTGATTGAAATGAACTCGA-CTCG------AG----------------------------------------------------------TGCCACAAA |
| droPer1 | super_10:2201269-2201389 - | GAGGAGAGCTG--TT---------------TCGCCGGACAGTCAGAGTTTGAGTTGATCTCTAGTTACTGGGCTTTGA--TTCGAAATAAGTAGCTTATTATTCTAGTGATTGAAATGAACTCGA-CTCG------AG----------------------------------------------------------TGCCACAAA |
| droWil1 | scaffold_180708:8625990-8626125 - | GTGTGAAACT-----------------------TG----AGTTTGAGTTTACATTGATCACTGGTTAGTGAGGTT-CAAAGTCAGACC---CAACTTCGTTATCTGGCGATCAA-ATAAACCTGG-CTTACA----AAAGCTATAATTGCTGAAATGGATACACGT-----------------------T------T---TCTAT |
| droVir3 | scaffold_12963:3615378-3615429 + | G---------------------------------------------------------------------------------------------TTTCATTTTTCAGTGAATTA-AACAACTCAA-TTTACACACACA----------------------------------------------------------CACACACAT |
| droMoj3 | scaffold_6500:14018312-14018360 + | ATTTAAATGTA--TT---------------CCGGTCTATGGCG--------------------TTTACGG--------------------------TCGTTTTGTCGTA---------------------------------------------------------------------------------------------CTC |
| droGri2 | scaffold_15252:3147112-3147237 + | GTTGTTAGCAG--CCACATCTCTGAATTGAAT-TG----GATTCGTT-----------------TTACT--------------------------TTCTTTTATTACTGAAATG-AGTTGTTCAG-TTC-T-TTTAAA-TT-------------------------TCAGTCTACAAACTAAGAGCAGGTTACTTTCATAACACA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641176-5641294 + | GTAGTGAGCTC--GTTATCAAG------------GGCCGAGTTACCTTTGATCACCAGTAACTGAGATT--GTTTCTGATA---CGGTTTCGTTTTCTGGCAATCA-AAAGAACTTGGACTCGACTTGAAT-T--CAA-----------------------CTAC |
| droSim1 | chr2L:5434262-5434381 + | GTAATGTGCTC--GTTATCATG------------GGCCGAGTTACCTTTGATCATCAGTAAGTGAGATT--GTTTCTGATA---CGATTTCGTTTTCTGGCAATCA-AAGGAACTCG-ACTCGACTTGAAT-CTACAA-----------------------CTAC |
| droSec1 | super_5:3718917-3719036 + | GTAATGTACTC--TTTATCAAG------------GCCCGAGTTACCTTTGATCACCAGTAAGTGAGATT--GTTTCTGATA---CGATTTCGTTTTCTGGCAATCA-AAGGAACTCG-ACTCGACTTGAAT-CTACAA-----------------------CTAC |
| droYak2 | chr2L:13552021-13552135 - | GAAAA----TCTAATT--CATG------------GGTCGAGTTACCATTGATCACCGGTTACTGAGGTT--GTTCCTTCTT---TGGCTTCGTTTTCTGGCGATCA-AAAGAACTCG-ACTCGACTCGAAT-C--CAA-----------------------CCAA |
| droEre2 | scaffold_4929:5729649-5729755 + | TG-----------ATT--CATG------------GGTCGAGTTACCATTGATCACCGGTTACTGAGGTT--GTGCCACATT---CGGCTTCGTTTCCTGGCGATCA-AAAGAACTCG-ACTCGGCTCGAAT-C--CCA-----------------------GTAC |
| droAna3 | scaffold_12916:11084979-11085099 + | ACAAAGGACTA--CA-----GGAAAGCGACTTAAGTCCGAGCACTATTTGATCACTGGTTACTGAGGTT--GTTTTGTCAT---AAGCTTCGTTTTCTGGCGATCA-AATACGCTCG-ACT----TTGGGC-C--CAA-----------------------CCAC |
| dp4 | chr4_group4:3182338-3182458 - | GAGGAGAGCTG--TTTCGCCGGA---------CAGTCAGAGTTTGATTTGATCTCTAGTTACTGGGCTTT-GATTCGAAATAAGTAGCTTATTATTCTAGTGATTGAAATGAACTCG-ACT-----CGAGT-G--CCA-----------------------CAAA |
| droPer1 | super_10:2201269-2201389 - | GAGGAGAGCTG--TTTCGCCGGA---------CAGTCAGAGTTTGAGTTGATCTCTAGTTACTGGGCTTT-GATTCGAAATAAGTAGCTTATTATTCTAGTGATTGAAATGAACTCG-ACT-----CGAGT-G--CCA-----------------------CAAA |
| droWil1 | scaffold_180708:8625990-8626125 - | GTGTGAAACTT----------G-----------AGTTTGAGTTTACATTGATCACTGGTTAGTGAGGTTCAAAGTCAGACC---CAACTTCGTTATCTGGCGATCA-AATAAACCTG-GCTTA---CAAAAGCTATAATTGCTGAAATGGATACACGTTTTCTAT |
| Species | Read pileup | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||
| dp4 |
|
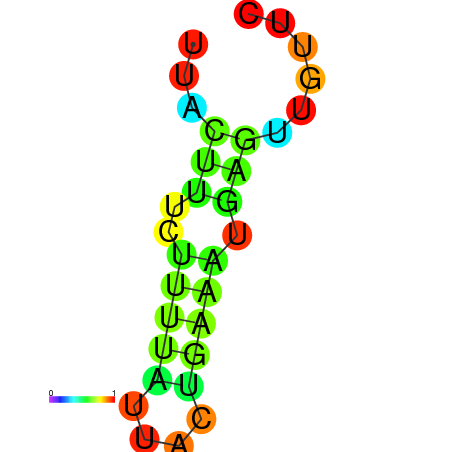
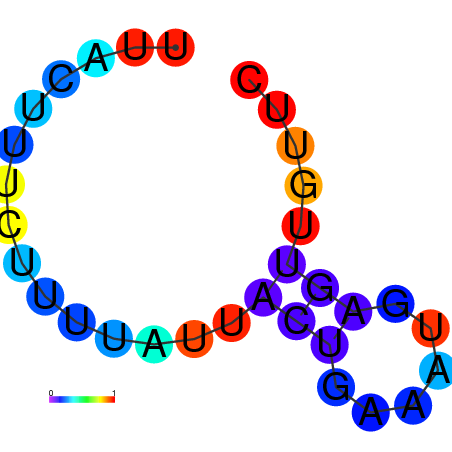
| dG=-17.9, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.0, p-value=0.009901 | dG=-17.0, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-23.0, p-value=0.009901 | dG=-22.6, p-value=0.009901 |
|---|---|
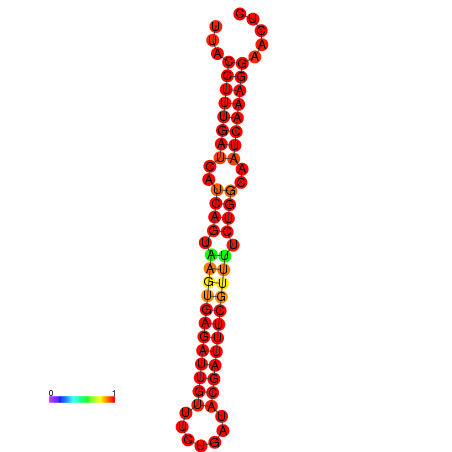 |
 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
 |
 |
| dG=-18.0, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.1, p-value=0.009901 | dG=-17.1, p-value=0.009901 |
|---|---|---|---|
 |
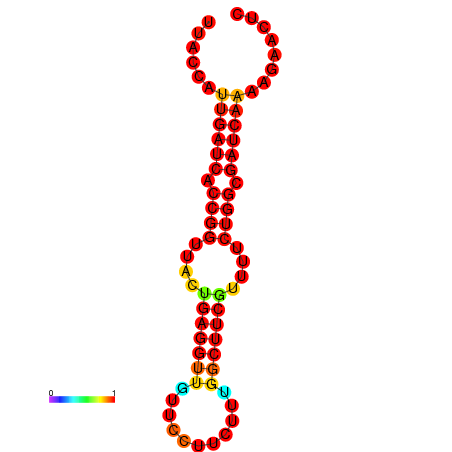 |
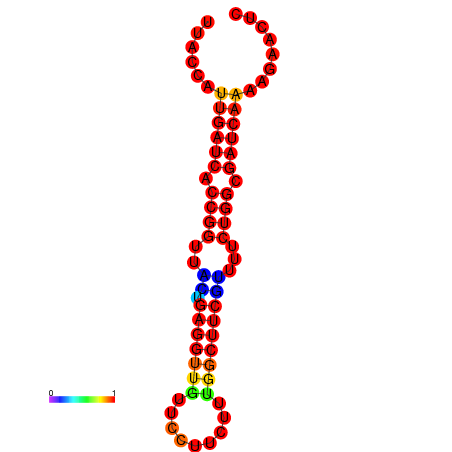 |
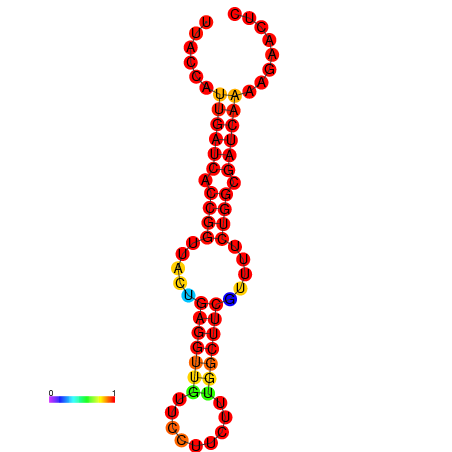 |
| dG=-20.2, p-value=0.009901 | dG=-19.7, p-value=0.009901 |
|---|---|
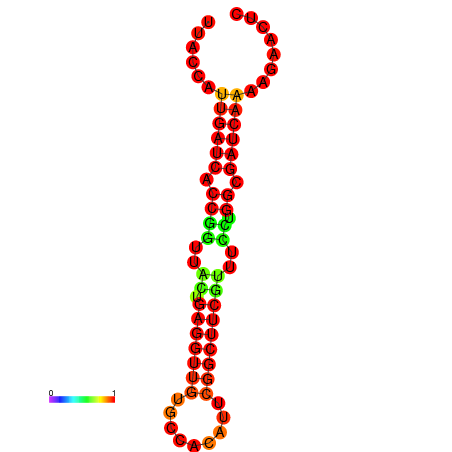 |
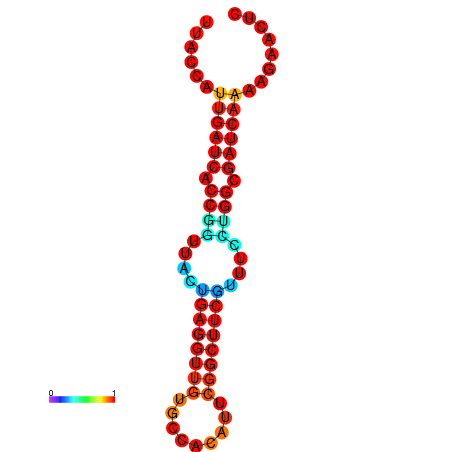 |
| dG=-18.7, p-value=0.009901 | dG=-17.8, p-value=0.009901 | dG=-17.8, p-value=0.009901 | dG=-17.7, p-value=0.009901 |
|---|---|---|---|
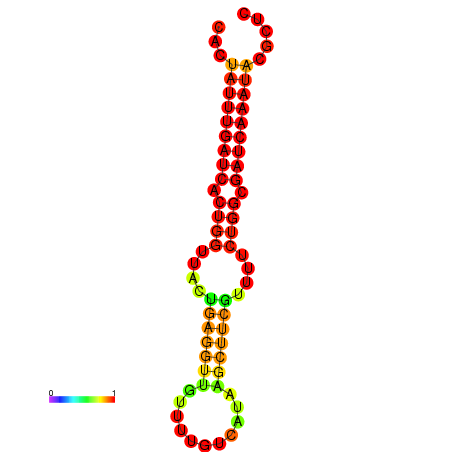 |
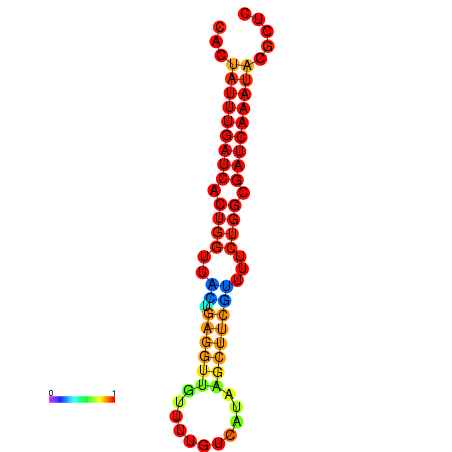 |
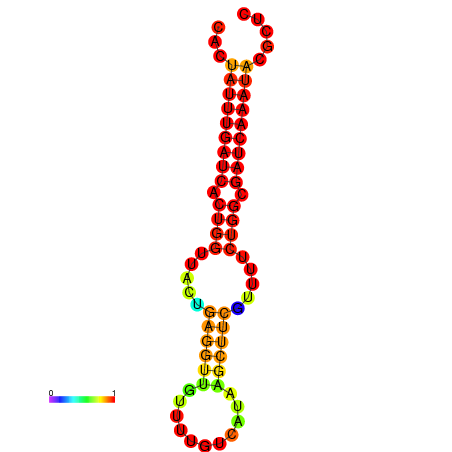 |
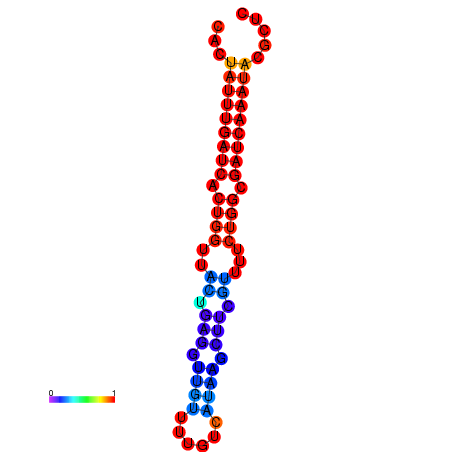 |
| dG=-13.0, p-value=0.009901 | dG=-12.7, p-value=0.009901 | dG=-12.5, p-value=0.009901 | dG=-12.4, p-value=0.009901 | dG=-12.2, p-value=0.009901 |
|---|---|---|---|---|
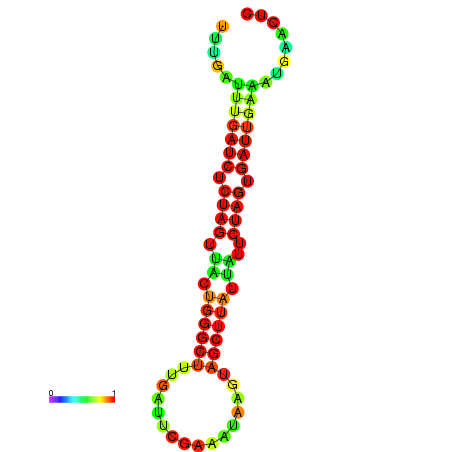 |
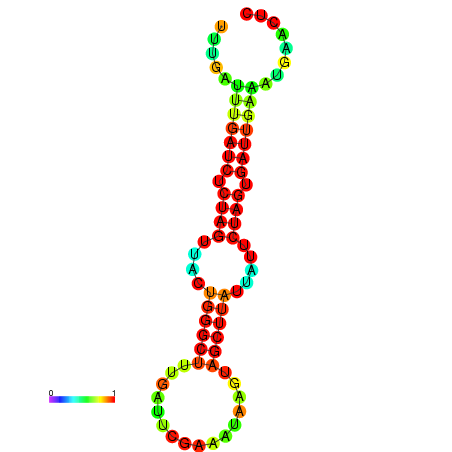 |
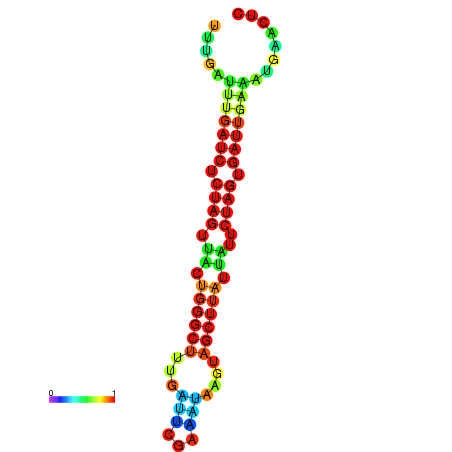 |
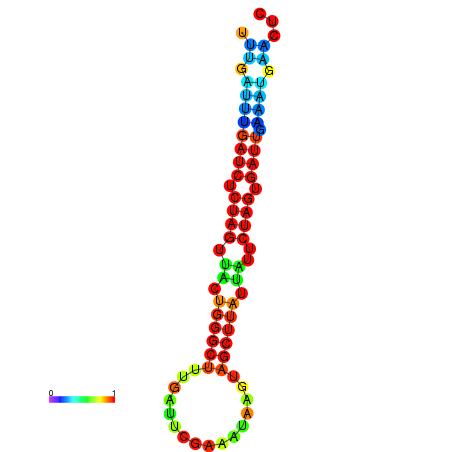 |
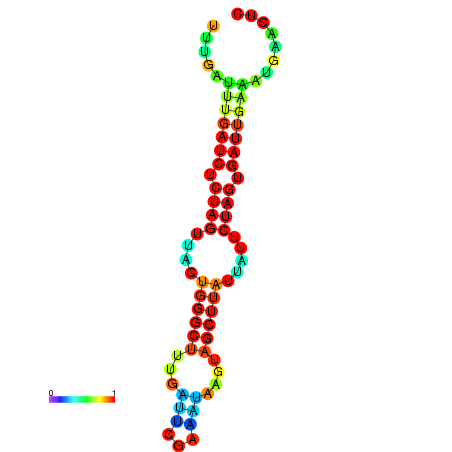 |
| dG=-15.2, p-value=0.009901 | dG=-15.1, p-value=0.009901 | dG=-14.7, p-value=0.009901 | dG=-14.4, p-value=0.009901 | dG=-14.3, p-value=0.009901 |
|---|---|---|---|---|
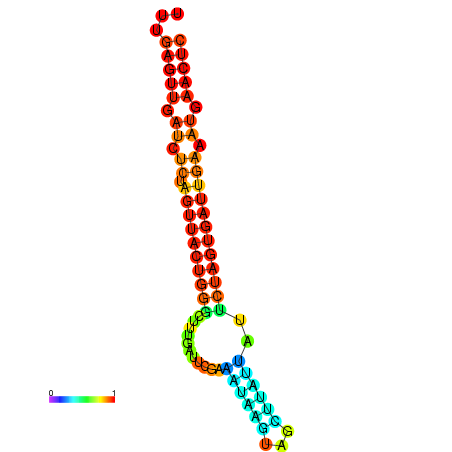 |
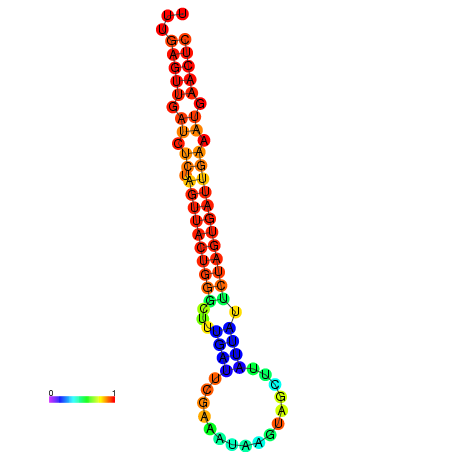 |
 |
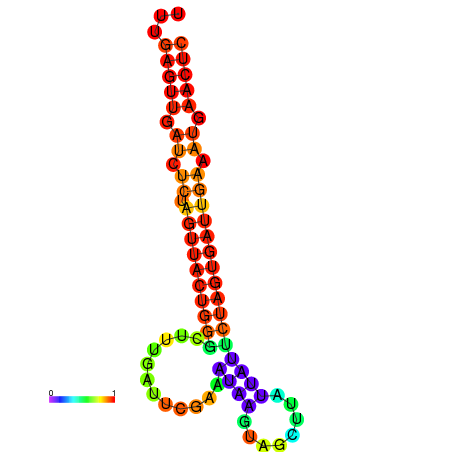 |
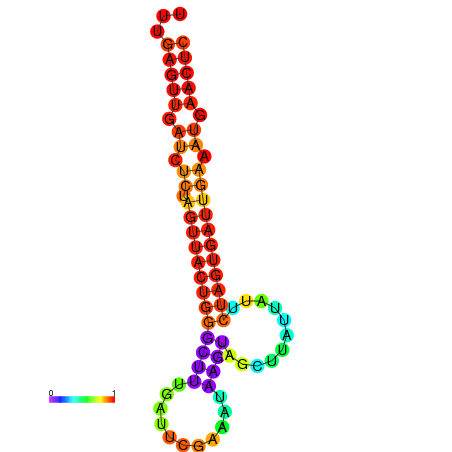 |
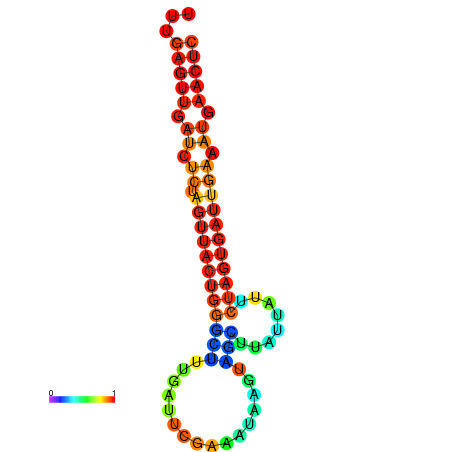
| dG=-16.4, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.5, p-value=0.009901 | dG=-15.5, p-value=0.009901 |
|---|---|---|---|
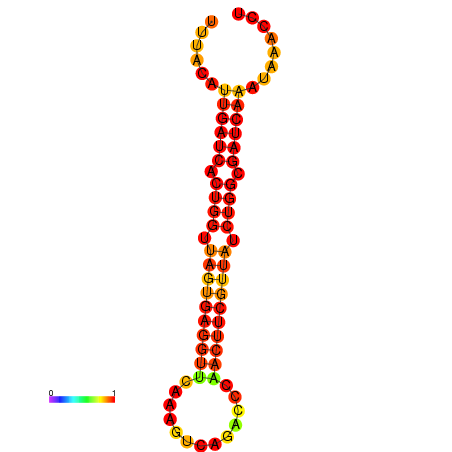 |
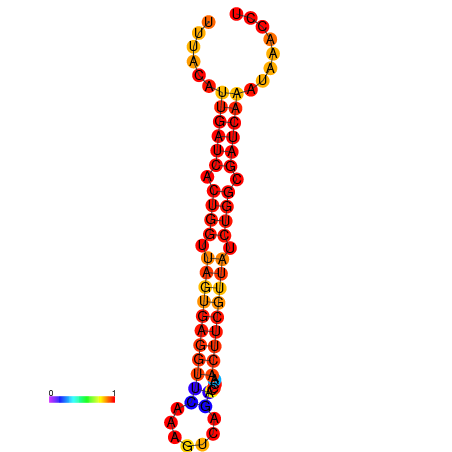 |
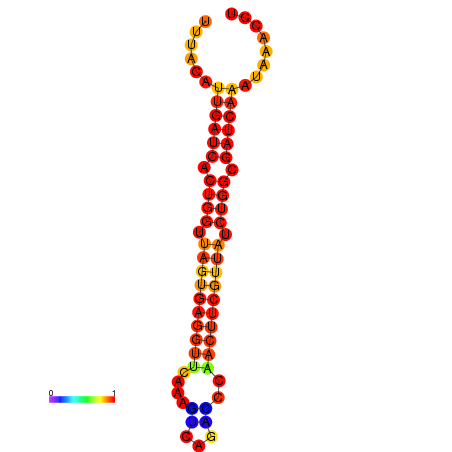 |
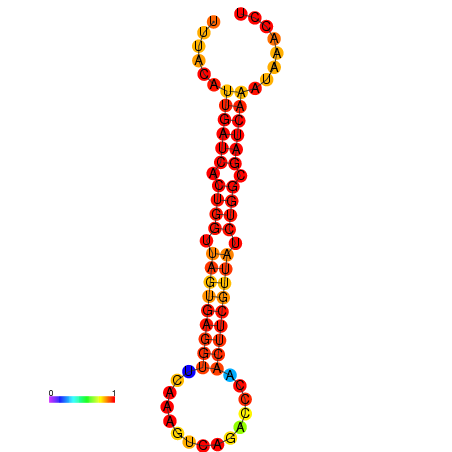 |
| dG=-1.9, p-value=0.009901 | dG=-1.6, p-value=0.009901 | dG=-1.2, p-value=0.009901 | dG=-0.9, p-value=0.009901 |
|---|---|---|---|
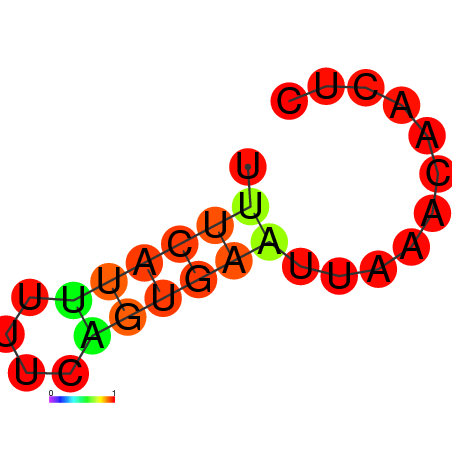 |
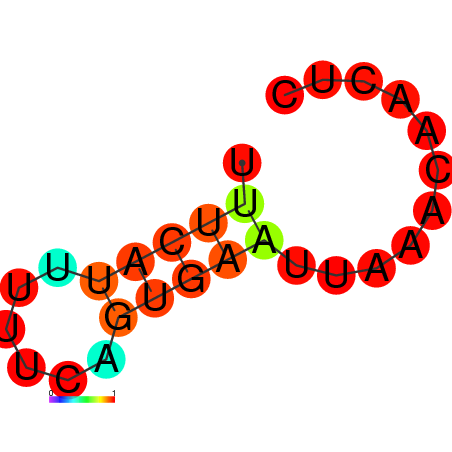 |
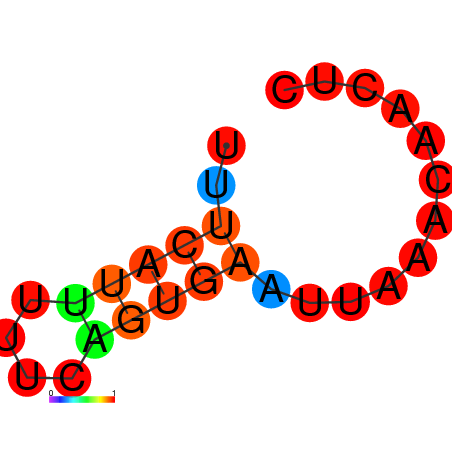 |
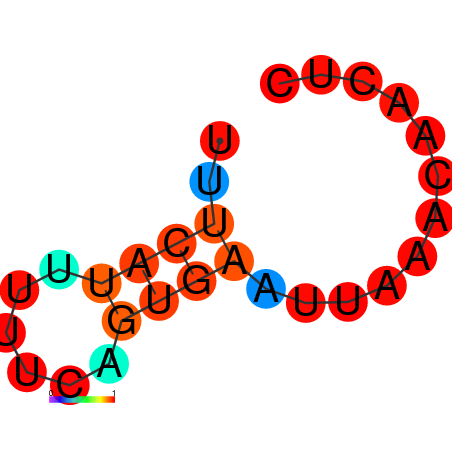 |
| dG=-1.7, p-value=0.009901 | dG=-1.3, p-value=0.009901 | dG=-0.8, p-value=0.009901 |
|---|---|---|
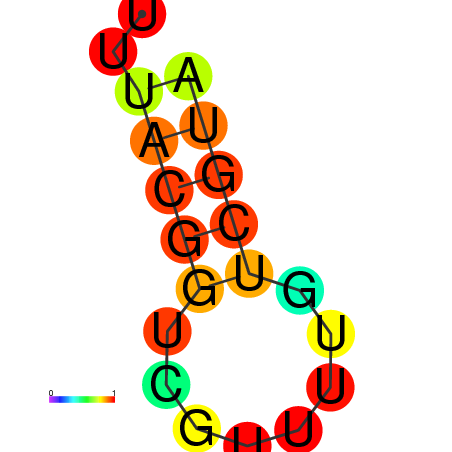 |
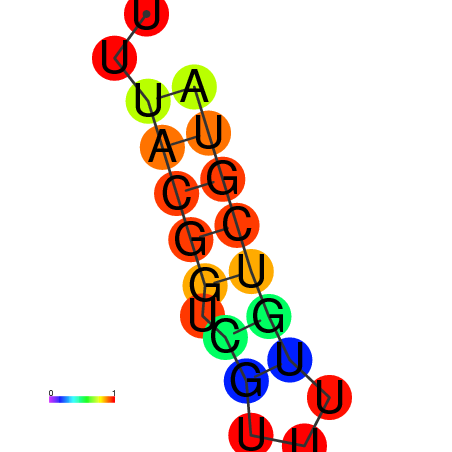 |
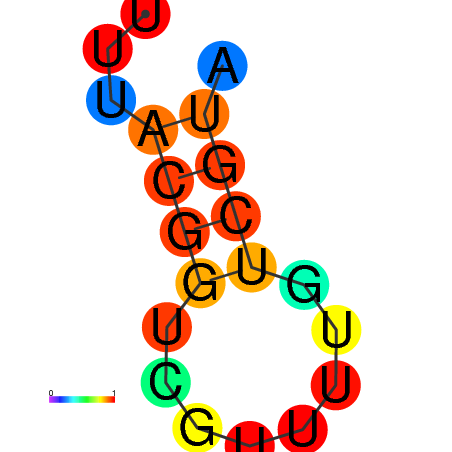 |
| dG=-1.7, p-value=0.009901 | dG=-0.9, p-value=0.029703 | dG=-0.7, p-value=0.009901 | dG=-0.7, p-value=0.009901 |
|---|---|---|---|
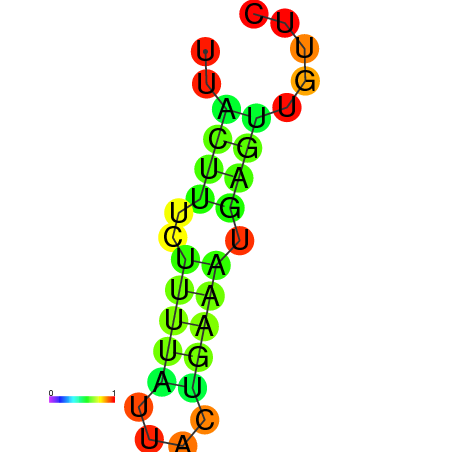 |
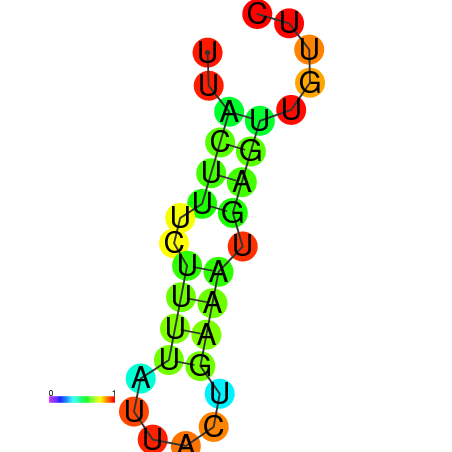 |
 |
 |
Generated: 03/07/2013 at 07:09 PM