| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641048-5641170 + | TATC---G-----TA------------CCGCCCAC---------------------G---GGTTTTGTACCACATTCTGAGTATTCCAGATTGC-ATAGCTTTGTGCTAC------TATTGCTATACGGTCTGGGACACTTTTAACATGGTATCAA-ATCAG---------ATTCA----------------CCTAATAG |
| droSim1 | chr2L:5434134-5434256 + | CATC---G-----TC------------ACGCCCAC---------------------G---GGTTTTGTACCACATTCAGAGTATTCCAGGTTGC-ATAGCTTTGTGCTAC------TATTGCTGTACGATCTGGGACACTTTTAACATGGTATCAA-ATCTG---------ATTCC----------------CCCAAACG |
| droSec1 | super_5:3718789-3718911 + | CATC---G-----TA------------CCGCCCAC---------------------G---GGTTTTGTACCACATTCTGAGTATTCCAGATTGC-ATAGCTTTGTGCTAC------TATTGCTGTACGATCTGGGAGACTTTTAACATGGTATCAA-ATCTG---------ATTCC----------------CCCAAAGG |
| droYak2 | chr2L:13552137-13552259 - | CATC---G-----CT------------TCGCCCGC---------------------G---TGTTTTGTACCAGATTCTGAGTATTCCTGATTGC-ATAGCTTTGGCCTTT------GATTGCTATACGGTCTGGGACACTTTTAACGTGGTATCAA-ATCTG---------ATTCA----------------ACCGAAGG |
| droEre2 | scaffold_4929:5729522-5729643 + | CAAC---G-----CA------------CCGCCCGC---------------------G---TTTTTTGTGCCAGATTCAGAGTATTCCAGATTGC-ATAGCTTTGGGCTTT------GATTGCTATACGGTCTGGGACACTTTAAACGTGGCATCAA--TCCT---------ATTCA----------------ACTGATGG |
| droAna3 | scaffold_12916:11084824-11084972 + | TATC---G-----GCCAATAGGCCAATTAACCCGGACC---A------ATCAATATG---AGTTTTGTGCCAGATTCTGAGTATTCCTGATTGC-ATAGCTCTGTGTTGT------GACTGCTATACGGTCTGGGACACTTCTAACATGGCATCAA-ATCTG---------TGTCAAC--------------GGCAGCAG |
| dp4 | chr4_group4:3182464-3182586 - | TGTG---G-----GT------------GTGCGTGT---------------------GTGCGGGTGTGTGCTATATTCTGAGTATTGCCTCCTGTTGTAGTTGTGGAATATTGAAATATCAACTATACGGTTCGGAATCCTCTAAATATGGCAGCC--------------------A----------------CTAAAGAC |
| droPer1 | super_10:2201395-2201525 - | TATACACGTAAATGT------------GTGTGGGT---------------------GTGCGGGTGTGTTCTATATTCTGAGTATTGCCTCCTGTTGTAGTTGTGGAATATTGAAATATCAACTATACGGTTCGGAATCCTCTAAATATGGCAGCC--------------------A----------------CTAAAGAC |
| droWil1 | scaffold_180708:8626131-8626297 - | TAAG---A-----AT------------TAACTTTGAATTGGAAACGATATTGAAA-T---GAATTTATGCTAAATTCTGAGTATTCCTGATTGC-ATAGTGTGTATTTGA------TACTACTATACGGTCGGGGAAACTTTTAACATGGCCCAAA--TGATAAATGACAGTGACAATGAAAAAGAAAATACCAAAACAG |
| droVir3 | Unknown | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 | scaffold_6500:14018206-14018306 + | TAA-----------------------------------------------------------------GTCATGCTTTGAGTATTCCTTATTGC-ATAGTTTAC--ATAT------AATTACTATACAGTCTGGAGCACTTAAAGTTGGGCATAGACAACCG---------AACAA----------------CTTAGGAG |
| droGri2 | scaffold_15252:3146997-3147109 + | TGTT---G-----AT------------TGGCCTTG---------------------G-------TTATGCTAAATTCCAAGTATTCCTTATTGC-ATAGTTTATAAT--T------GAATACTATACAATCTGGCGTTCTTAGAATTTGTCATAAG-AC-------------TTCA----------------CATAACAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5641048-5641170 + | TATC---G-----TA------------CCGCCCAC---------------------G---GGTTTTGTACCACATTCTGAGTATTCCAGATTGC-ATAGCTTTGTGCTAC------TATTGCTATACGGTCTGGGACACTTTTAACATGGTATCAA-ATCAG---------ATTCA----------------CCTAATAG |
| droSim1 | chr2L:5434134-5434256 + | CATC---G-----TC------------ACGCCCAC---------------------G---GGTTTTGTACCACATTCAGAGTATTCCAGGTTGC-ATAGCTTTGTGCTAC------TATTGCTGTACGATCTGGGACACTTTTAACATGGTATCAA-ATCTG---------ATTCC----------------CCCAAACG |
| droSec1 | super_5:3718789-3718911 + | CATC---G-----TA------------CCGCCCAC---------------------G---GGTTTTGTACCACATTCTGAGTATTCCAGATTGC-ATAGCTTTGTGCTAC------TATTGCTGTACGATCTGGGAGACTTTTAACATGGTATCAA-ATCTG---------ATTCC----------------CCCAAAGG |
| droYak2 | chr2L:13552137-13552259 - | CATC---G-----CT------------TCGCCCGC---------------------G---TGTTTTGTACCAGATTCTGAGTATTCCTGATTGC-ATAGCTTTGGCCTTT------GATTGCTATACGGTCTGGGACACTTTTAACGTGGTATCAA-ATCTG---------ATTCA----------------ACCGAAGG |
| droEre2 | scaffold_4929:5729522-5729643 + | CAAC---G-----CA------------CCGCCCGC---------------------G---TTTTTTGTGCCAGATTCAGAGTATTCCAGATTGC-ATAGCTTTGGGCTTT------GATTGCTATACGGTCTGGGACACTTTAAACGTGGCATCAA--TCCT---------ATTCA----------------ACTGATGG |
| droAna3 | scaffold_12916:11084824-11084972 + | TATC---G-----GCCAATAGGCCAATTAACCCGGACC---A------ATCAATATG---AGTTTTGTGCCAGATTCTGAGTATTCCTGATTGC-ATAGCTCTGTGTTGT------GACTGCTATACGGTCTGGGACACTTCTAACATGGCATCAA-ATCTG---------TGTCAAC--------------GGCAGCAG |
| dp4 | chr4_group4:3182464-3182586 - | TGTG---G-----GT------------GTGCGTGT---------------------GTGCGGGTGTGTGCTATATTCTGAGTATTGCCTCCTGTTGTAGTTGTGGAATATTGAAATATCAACTATACGGTTCGGAATCCTCTAAATATGGCAGCC--------------------A----------------CTAAAGAC |
| droPer1 | super_10:2201395-2201525 - | TATACACGTAAATGT------------GTGTGGGT---------------------GTGCGGGTGTGTTCTATATTCTGAGTATTGCCTCCTGTTGTAGTTGTGGAATATTGAAATATCAACTATACGGTTCGGAATCCTCTAAATATGGCAGCC--------------------A----------------CTAAAGAC |
| droWil1 | scaffold_180708:8626131-8626297 - | TAAG---A-----AT------------TAACTTTGAATTGGAAACGATATTGAAA-T---GAATTTATGCTAAATTCTGAGTATTCCTGATTGC-ATAGTGTGTATTTGA------TACTACTATACGGTCGGGGAAACTTTTAACATGGCCCAAA--TGATAAATGACAGTGACAATGAAAAAGAAAATACCAAAACAG |
| droMoj3 | scaffold_6500:14018206-14018306 + | TAA-----------------------------------------------------------------GTCATGCTTTGAGTATTCCTTATTGC-ATAGTTTAC--ATAT------AATTACTATACAGTCTGGAGCACTTAAAGTTGGGCATAGACAACCG---------AACAA----------------CTTAGGAG |
| droGri2 | scaffold_15252:3146997-3147109 + | TGTT---G-----AT------------TGGCCTTG---------------------G-------TTATGCTAAATTCCAAGTATTCCTTATTGC-ATAGTTTATAAT--T------GAATACTATACAATCTGGCGTTCTTAGAATTTGTCATAAG-AC-------------TTCA----------------CATAACAA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-22.3, p-value=0.009901 | dG=-21.7, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.3, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-22.9, p-value=0.009901 | dG=-22.6, p-value=0.009901 | dG=-22.3, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 |
|---|---|---|---|---|
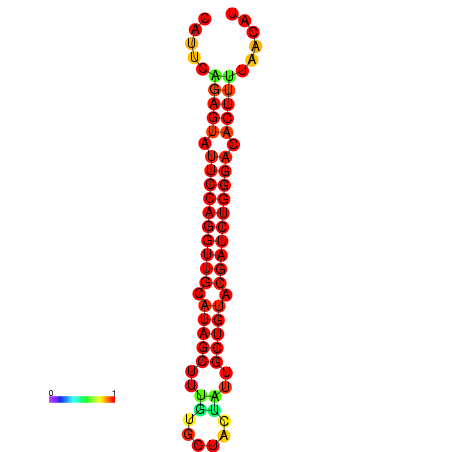 |
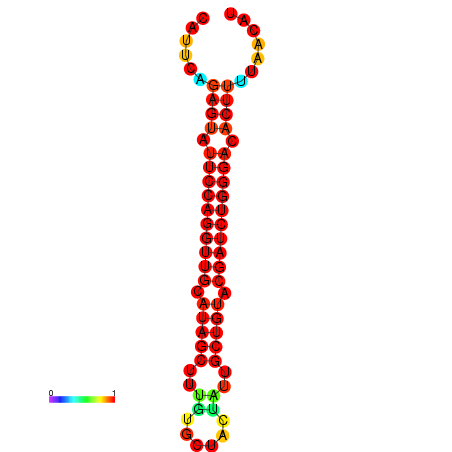 |
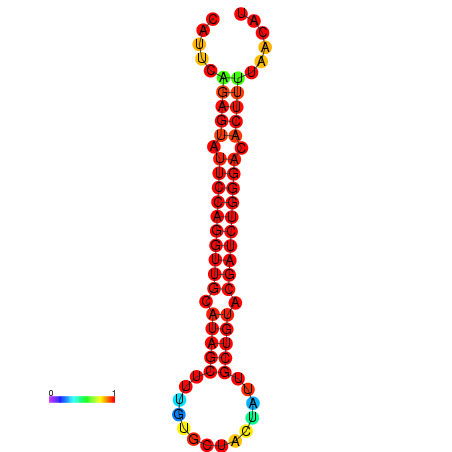 |
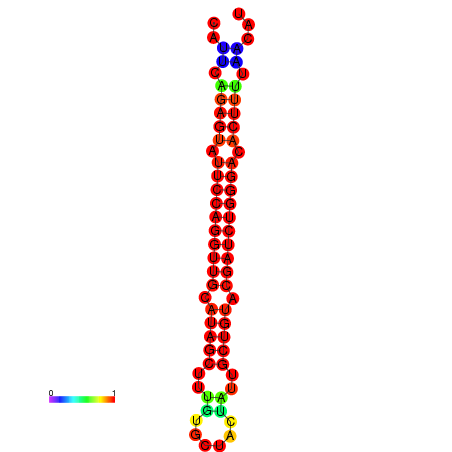 |
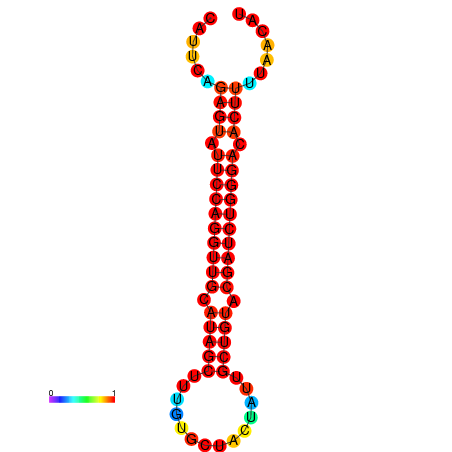 |
| dG=-22.8, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.8, p-value=0.009901 |
|---|---|---|---|---|
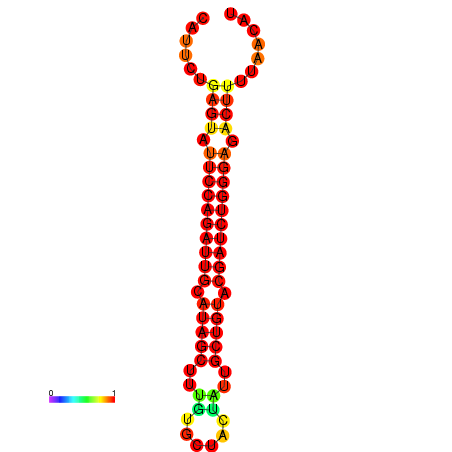 |
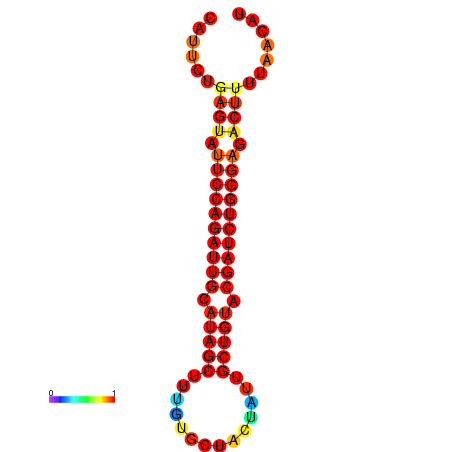 |
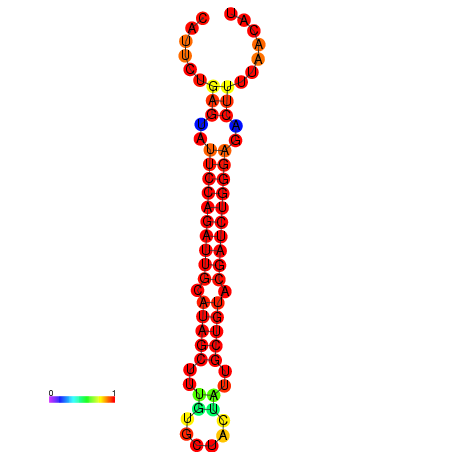 |
 |
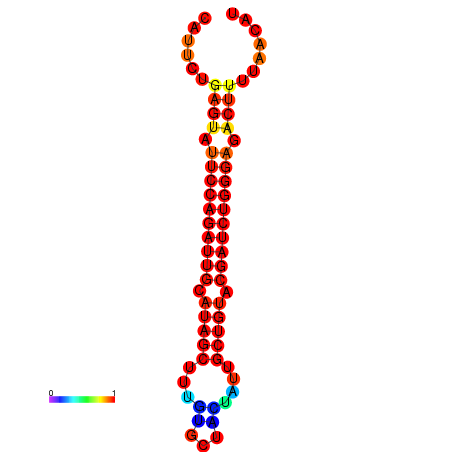 |
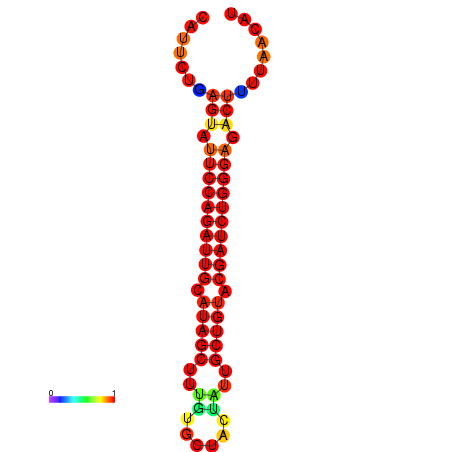
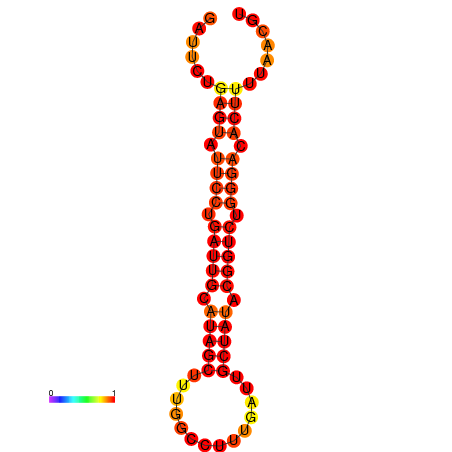
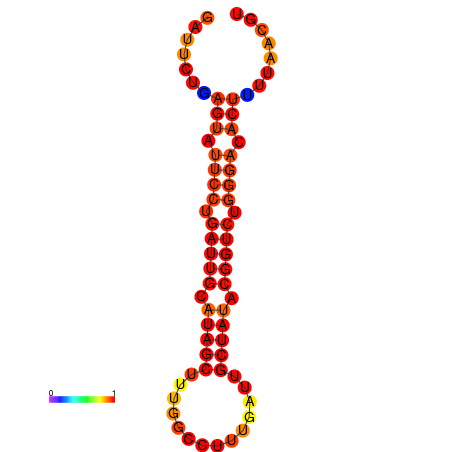
| dG=-17.1, p-value=0.009901 | dG=-16.2, p-value=0.009901 |
|---|---|
 |
 |
| dG=-22.3, p-value=0.009901 | dG=-22.1, p-value=0.009901 | dG=-21.5, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-18.1, p-value=0.009901 | dG=-17.8, p-value=0.009901 | dG=-17.3, p-value=0.009901 |
|---|---|---|
 |
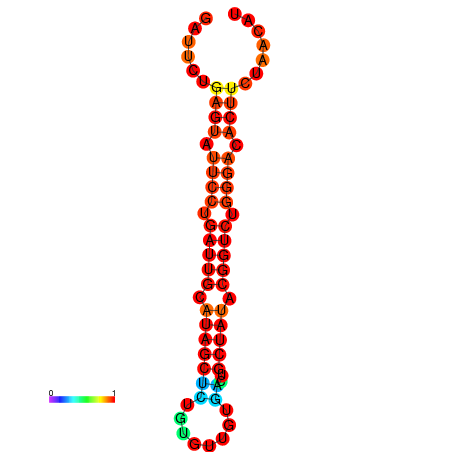 |
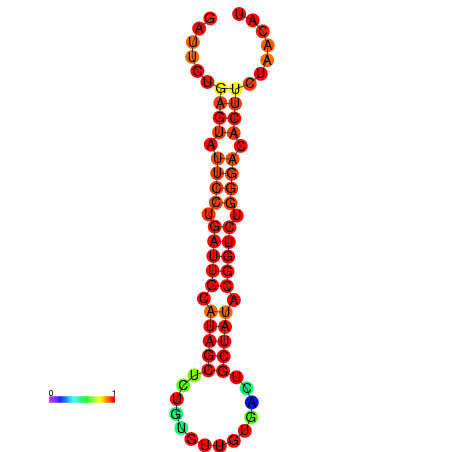 |
| dG=-14.3, p-value=0.009901 | dG=-14.3, p-value=0.009901 | dG=-13.7, p-value=0.009901 | dG=-13.7, p-value=0.009901 | dG=-13.5, p-value=0.009901 |
|---|---|---|---|---|
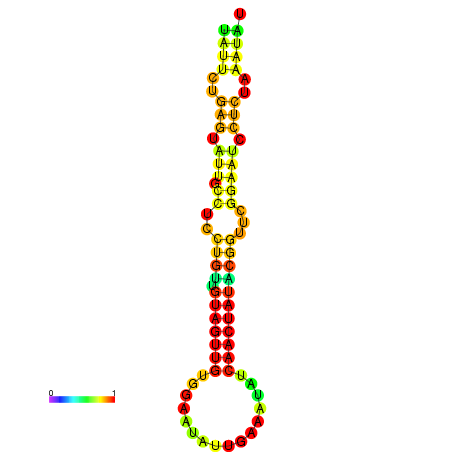 |
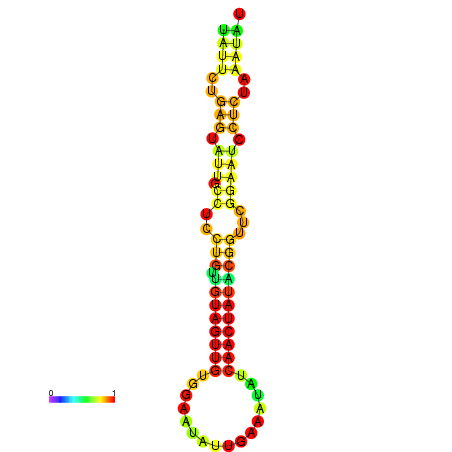 |
 |
 |
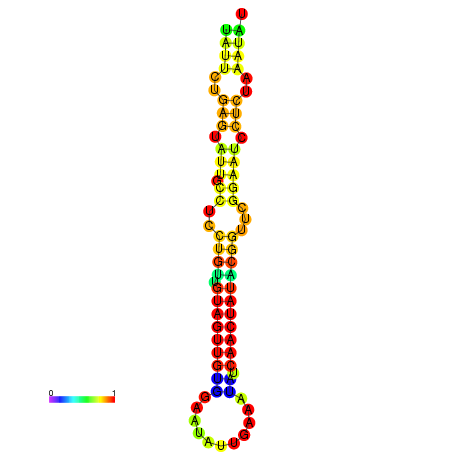 |
| dG=-14.3, p-value=0.009901 | dG=-14.3, p-value=0.009901 | dG=-13.7, p-value=0.009901 | dG=-13.7, p-value=0.009901 | dG=-13.5, p-value=0.009901 |
|---|---|---|---|---|
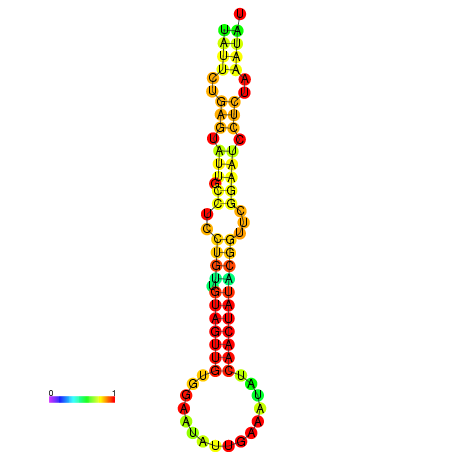 |
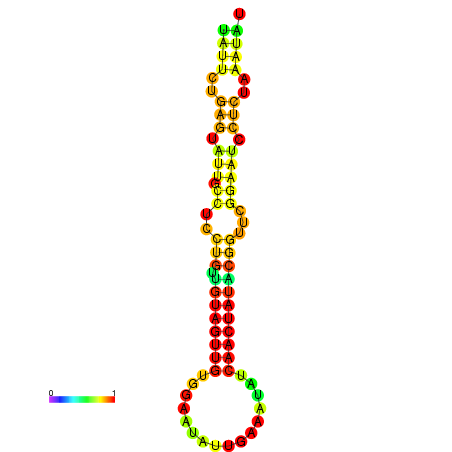 |
 |
 |
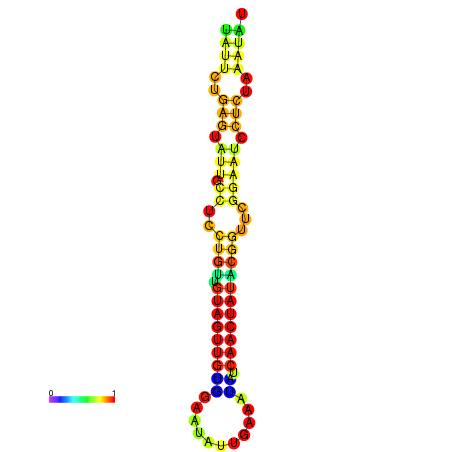 |
| dG=-22.6, p-value=0.009901 | dG=-22.3, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-21.7, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
No secondary structure predictions returned
| dG=-20.4, p-value=0.009901 |
|---|
 |
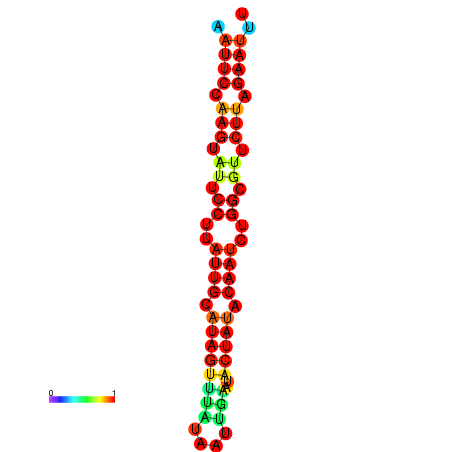
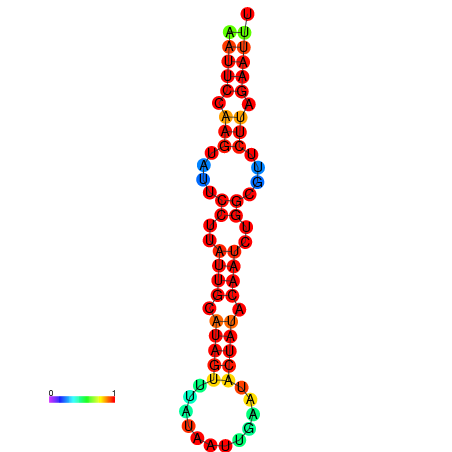
| dG=-12.4, p-value=0.009901 | dG=-12.3, p-value=0.009901 | dG=-11.9, p-value=0.009901 | dG=-11.8, p-value=0.009901 | dG=-11.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 07:08 PM