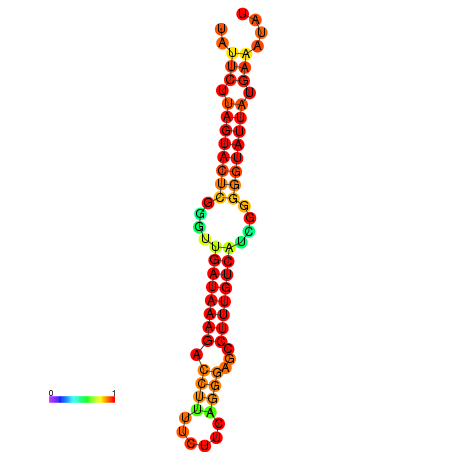
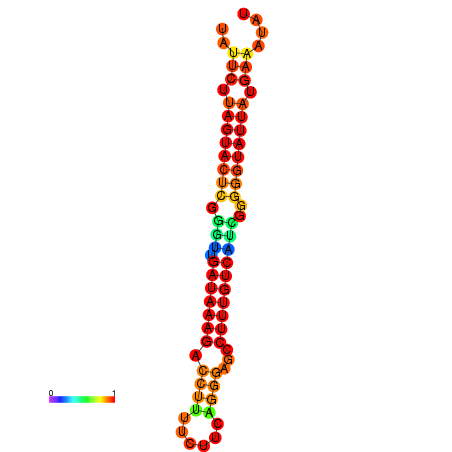
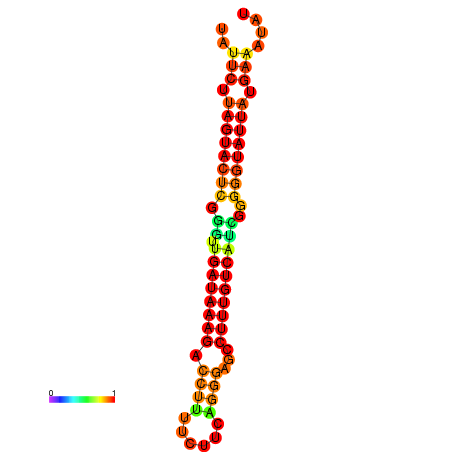

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5640925-5641052 + | CGAATTAA-ATTGATTTTAACTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTTCT---TCAG--GGAGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGTTTAAAGAAACTGCAATGATT------------ATCG---------------------------------- |
| droSim1 | chr2L:5434012-5434138 + | A--ATTAA-ATAGATTTTAACTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTTCT---TTGC--GGAGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGTTTAAAGAAGCTGCAAGGATC------------ATCG---------------------------------- |
| droSec1 | super_5:3718667-3718793 + | A--ATTAA-ATAGATTTTAACTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTTCT---TTGC--GGAGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGTTTAAAGAAGCTGCAAGGATC------------ATCG---------------------------------- |
| droYak2 | chr2L:13552255-13552381 - | CGAGTTAA-ATTGATTTTAACTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGACAAAGACCTTTCCT----TGC--GGCGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGCTCAAAGAAACTGCAAGGATC------------ATCG---------------------------------- |
| droEre2 | scaffold_4929:5729400-5729526 + | CGAGTTAA-ATTGATTTTAACTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGACAAAGAGCTTTCCT----TGC--GGCGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGCTCAAAGAAACTGCAAGGAGC------------AACG---------------------------------- |
| droAna3 | scaffold_12916:11084702-11084849 + | GAAGTTAA-ATTTATTTCAATTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTCCT----GGA--GCCGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGCTCAAAAAACCAGCACAATGT------------ATCGGCCAATAGGCCAATTAACCCG------------- |
| dp4 | chr4_group4:3182582-3182745 - | TGGTTTTC-GTTTA------------------------CGTTTTTT-TTTT--------GTTTTGGTTCTATGTTCTTAGTACTTGGGTTGATAAAGACCACTCGTAGTGTGA--GAAGCCTTTGTCATCGCGGGTATTATGCAACA---------TAGCTCAAAAGCGCAACAAAATACACA-------------AAA--------AATATACACGTAAATGTGTGTGG |
| droPer1 | super_10:2201518-2201669 - | TGGTTTTC-GTTCA------------------------CGTTTTTTTTTTT--------GTTTTGGTTCTATGTTCTTAGTACTTGGGTTGATAAAGACCACTCGTAGTGTGA--GAAGCCTTTGTCATCGCGGGTATTATGCAACA---------TAGCTCAAAAGCGCAACAAAATACACA-------------AAA--------AATATACACG------------- |
| droWil1 | scaffold_180708:8626283-8626424 - | AGTGAAAA-AGTGACTTAAGTTTT-----------------------------------------GTTCTATATTCTTAGTACTCGGGTTGATAAAGACAAAATGT---TTACCAATCGCCTTTGTCATCGGGGGTATTATGAAATA---------TAGCTCAAAAGTTCAACATAATAATTAAGAATTAACTTTG---------------------------------- |
| droVir3 | Unknown | -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 | scaffold_6500:14018136-14018207 + | TGATGTAATAAGTGTTTTAGCTTT-----------------------------------------AAGTTGTGCCATT-------------------------------------------------------------TTAAATTG---------AAAAAAGTG-AAGATTGAAAATTG------------ACTA---------------------------------- |
| droGri2 | scaffold_15252:3146904-3147001 + | CAAAGAGT-AGCAGGAGTAGTTAAGTTGTAGTTTCTTACGGTTTTTTTTTTAATAACATGCT--------------TTAGTGCT--------------------------------------------------------------TCAATGTAGGTGGCTCAA---------GATATTT------------GTTG---------------------------------- |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2L:5640925-5641052 + | CGAATTAAATTGA---------TTTTAACTTTGTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTTCT---TCAG--GGAGCCTTTGTCATCGGGGGTATTATGAAATATAGTTTAAAGAAACTGCAATGATTATCG----------------------------------- |
| droSim1 | chr2L:5434012-5434138 + | A--ATTAAATAGA---------TTTTAACTTTGTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTTCT---TTGC--GGAGCCTTTGTCATCGGGGGTATTATGAAATATAGTTTAAAGAAGCTGCAAGGATCATCG----------------------------------- |
| droSec1 | super_5:3718667-3718793 + | A--ATTAAATAGA---------TTTTAACTTTGTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTTCT---TTGC--GGAGCCTTTGTCATCGGGGGTATTATGAAATATAGTTTAAAGAAGCTGCAAGGATCATCG----------------------------------- |
| droYak2 | chr2L:13552255-13552381 - | CGAGTTAAATTGA---------TTTTAACTTTGTTCTATATTCTTAGTACTCGGGTTGACAAAGACCTTTCCT----TGC--GGCGCCTTTGTCATCGGGGGTATTATGAAATATAGCTCAAAGAAACTGCAAGGATCATCG----------------------------------- |
| droEre2 | scaffold_4929:5729400-5729526 + | CGAGTTAAATTGA---------TTTTAACTTTGTTCTATATTCTTAGTACTCGGGTTGACAAAGAGCTTTCCT----TGC--GGCGCCTTTGTCATCGGGGGTATTATGAAATATAGCTCAAAGAAACTGCAAGGAGCAACG----------------------------------- |
| droAna3 | scaffold_12916:11084702-11084849 + | GAAGTTAAATTTA---------TTTCAATTTTGTTCTATATTCTTAGTACTCGGGTTGATAAAGACCTTTCCT----GGA--GCCGCCTTTGTCATCGGGGGTATTATGAAATATAGCTCAAAAAACCAGCACAATGTATCGGCCAATAGGCCAATTA-ACCCG------------- |
| dp4 | chr4_group4:3182582-3182745 - | TGGTTTTCGTTTACGTTTTT-TTTTTGTTTTGGTTCTATGTTCTTAGTACTTGGGTTGATAAAGACCACTCGTAGTGTGA--GAAGCCTTTGTCATCGCGGGTATTATGCAACATAGCTCAAAAGCGCAACAAAATACACA-AAA--------AA-TATACACGTAAATGTGTGTGG |
| droPer1 | super_10:2201518-2201669 - | TGGTTTTCGTTCACGTTTTTTTTTTTGTTTTGGTTCTATGTTCTTAGTACTTGGGTTGATAAAGACCACTCGTAGTGTGA--GAAGCCTTTGTCATCGCGGGTATTATGCAACATAGCTCAAAAGCGCAACAAAATACACA-AAA--------AA-TATACACG------------- |
| droWil1 | scaffold_180708:8626283-8626424 - | AGTGAAAAAGTGA---------CTTAAGTTTTGTTCTATATTCTTAGTACTCGGGTTGATAAAGACAAAATGT---TTACCAATCGCCTTTGTCATCGGGGGTATTATGAAATATAGCTCAAAAGTTCAACATAATAATTA-AGA--------AT-TAACTTTG------------- |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-21.2, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.6, p-value=0.009901 | dG=-20.6, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-18.7, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|---|---|---|
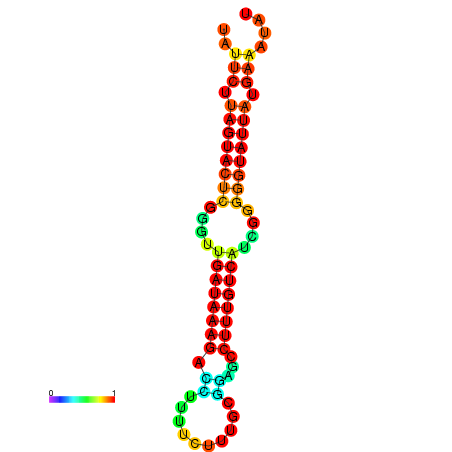 |
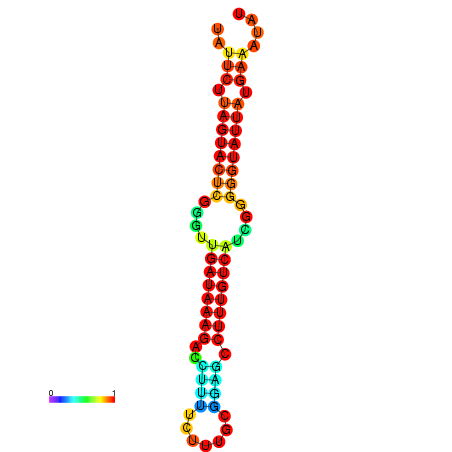 |
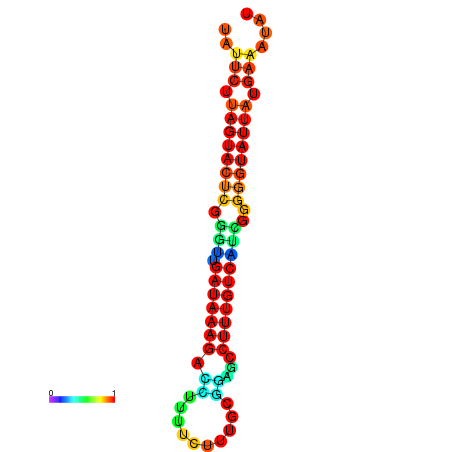 |
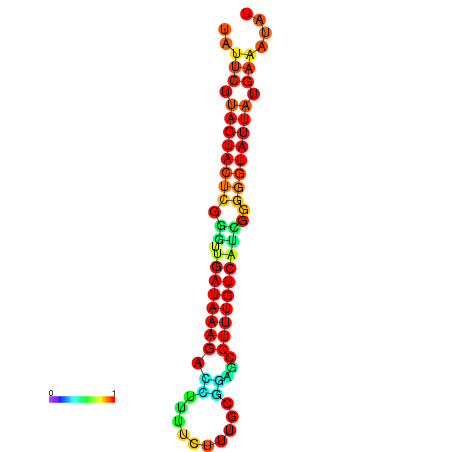 |
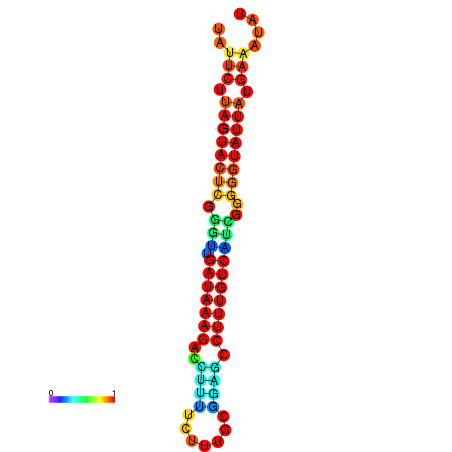 |
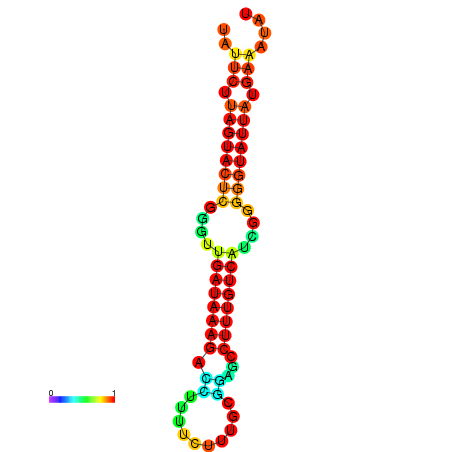
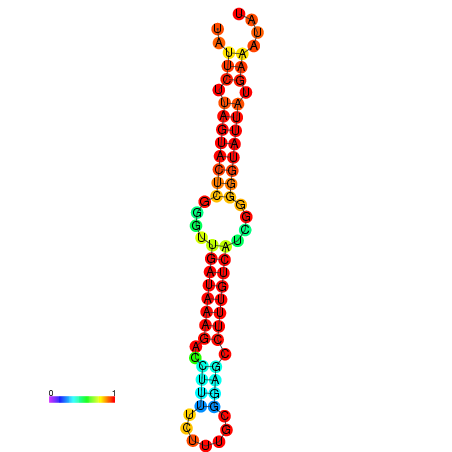
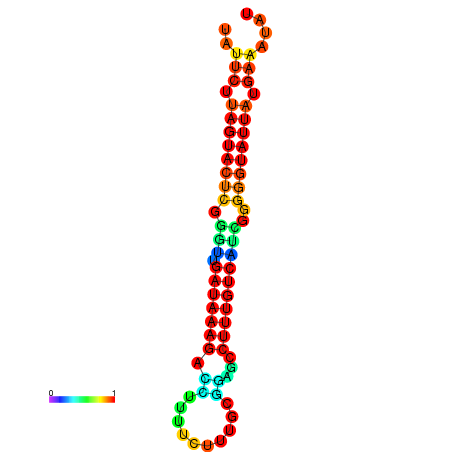
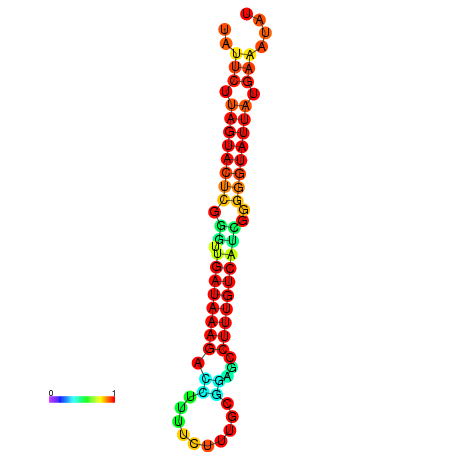
| dG=-18.7, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-18.1, p-value=0.009901 | dG=-17.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-20.7, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 |
|---|---|---|
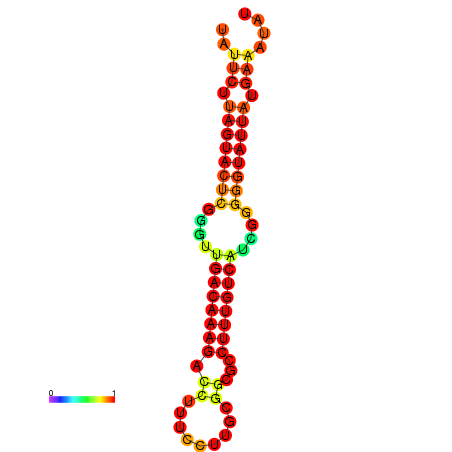 |
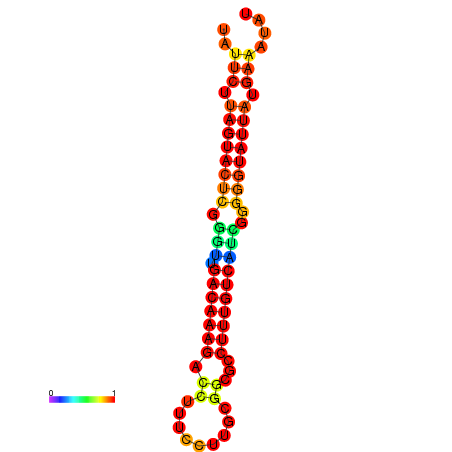 |
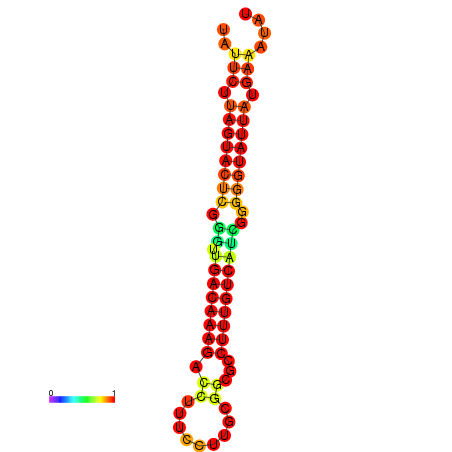 |
| dG=-24.4, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.7, p-value=0.009901 |
|---|---|---|---|
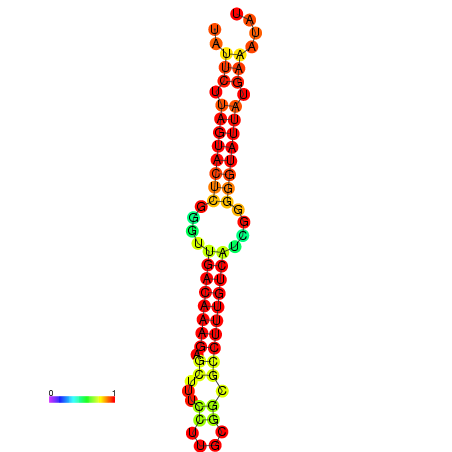 |
 |
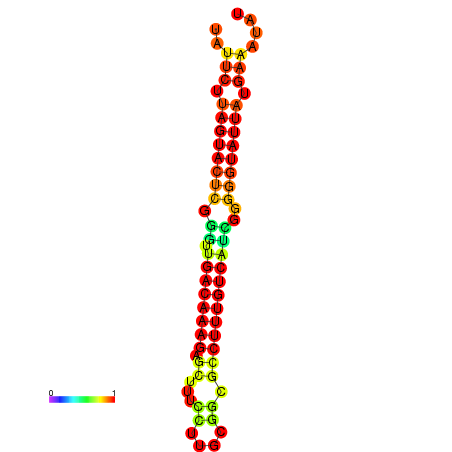 |
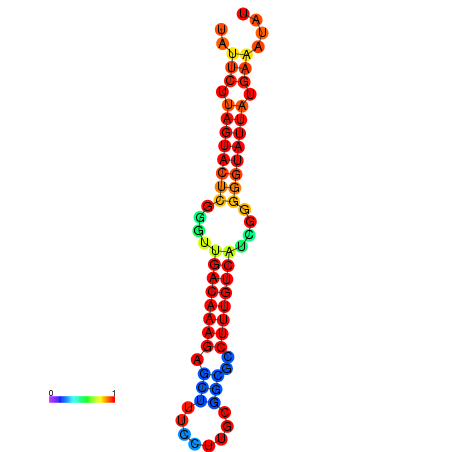 |
| dG=-17.7, p-value=0.009901 | dG=-17.6, p-value=0.009901 | dG=-17.5, p-value=0.009901 | dG=-17.4, p-value=0.009901 | dG=-17.2, p-value=0.009901 |
|---|---|---|---|---|
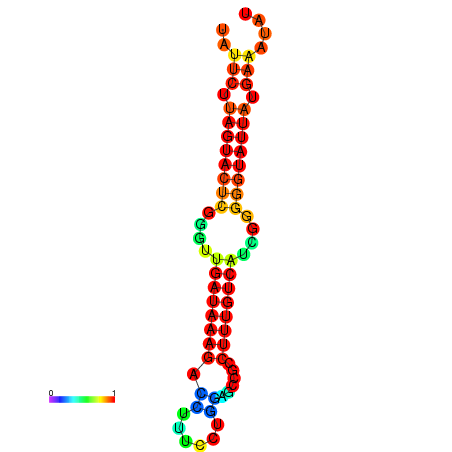 |
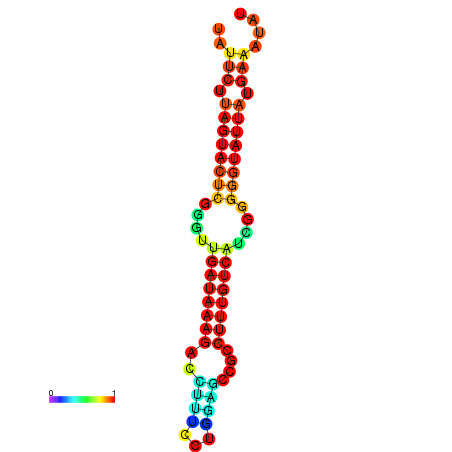 |
 |
 |
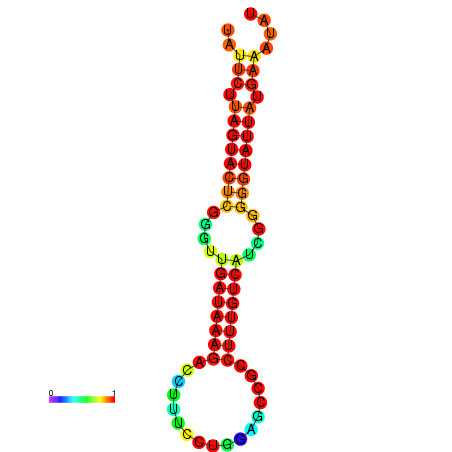 |
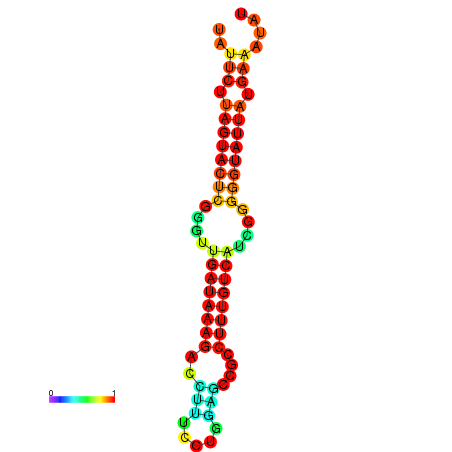
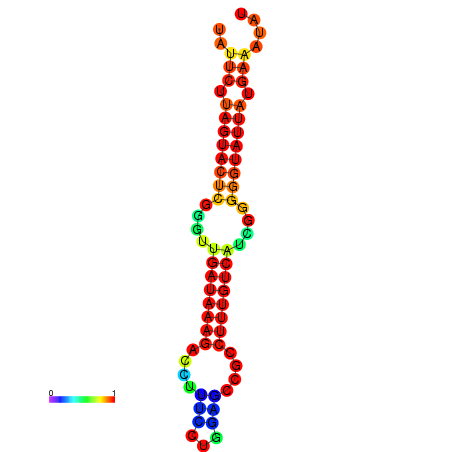
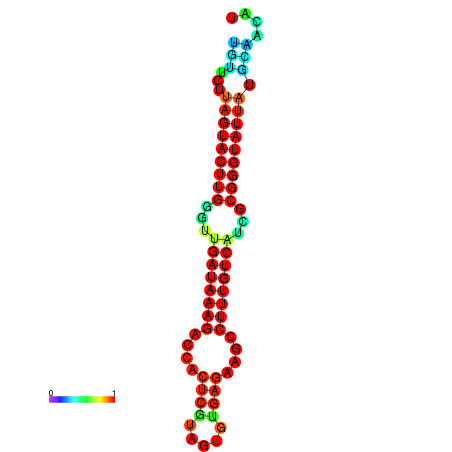
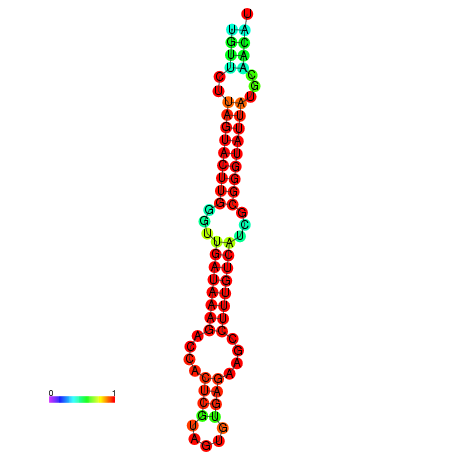
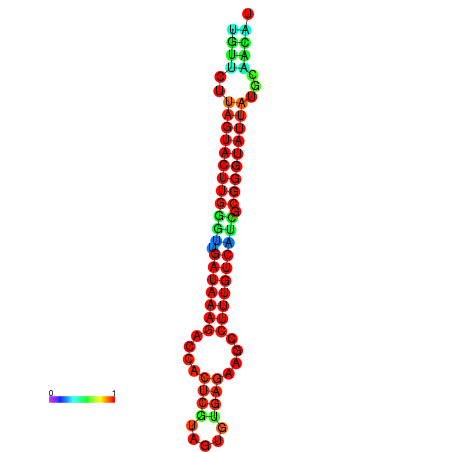
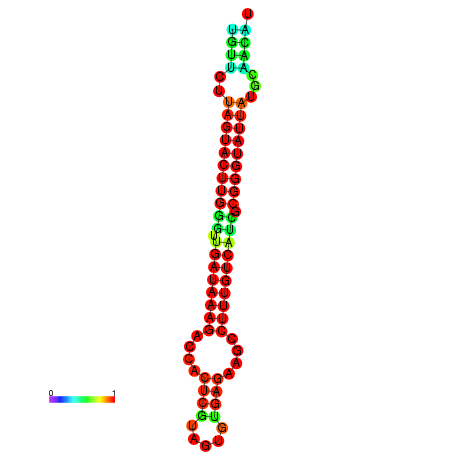
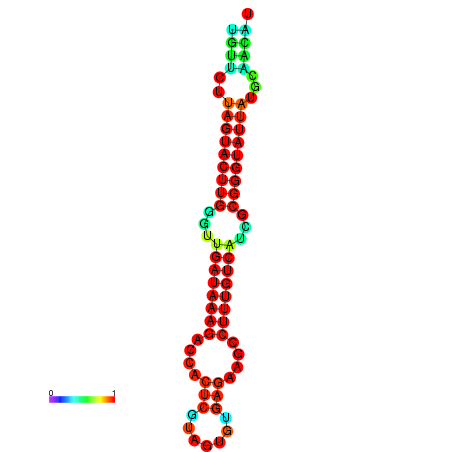
| dG=-19.6, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-19.6, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.3, p-value=0.009901 |
|---|---|---|---|---|
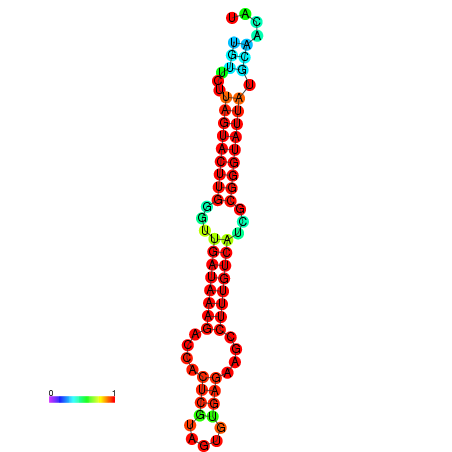 |
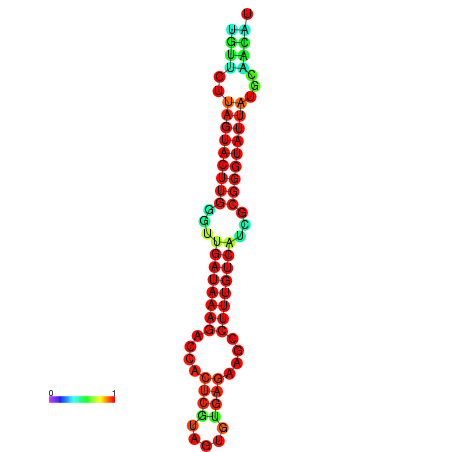 |
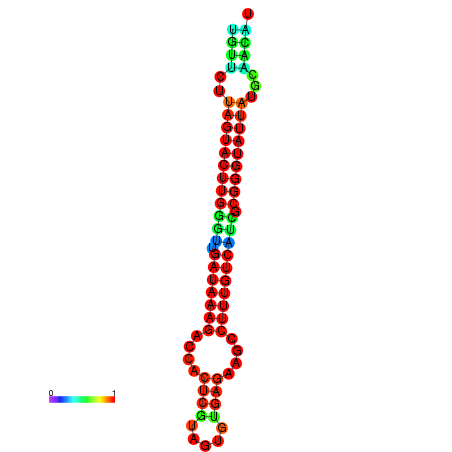 |
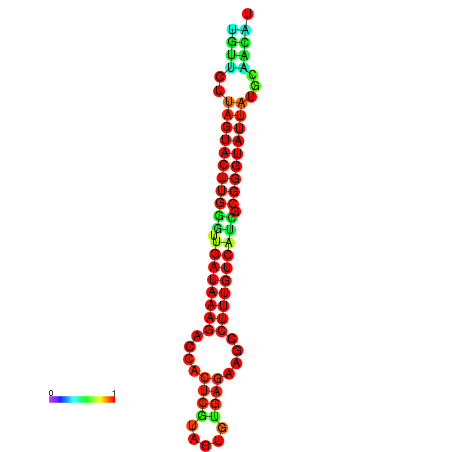 |
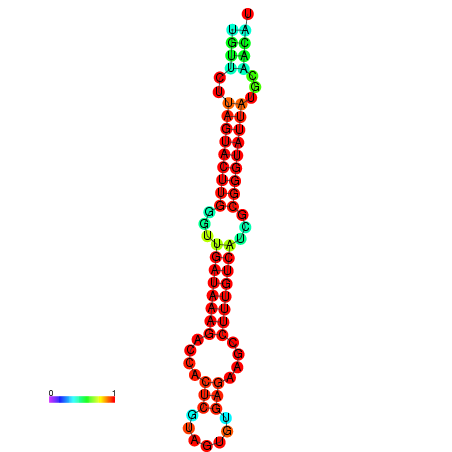 |
| dG=-17.1, p-value=0.009901 | dG=-16.5, p-value=0.009901 | dG=-16.5, p-value=0.009901 | dG=-16.3, p-value=0.009901 |
|---|---|---|---|
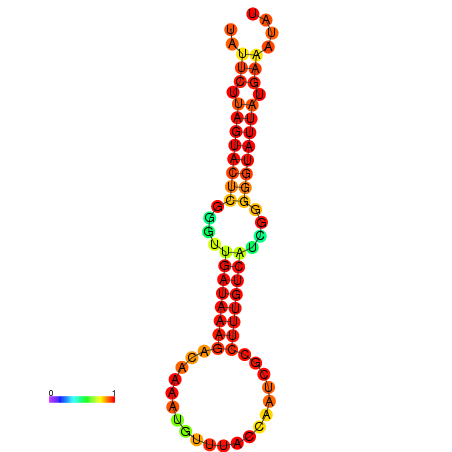 |
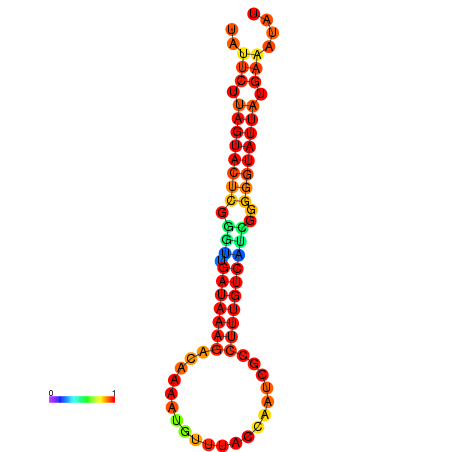 |
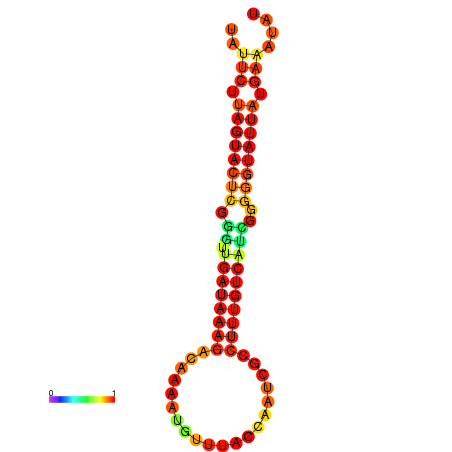 |
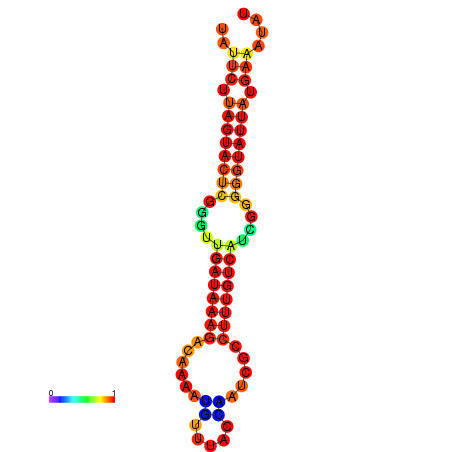 |
No secondary structure predictions returned
| dG=0.0, p-value=1.0 |
|---|
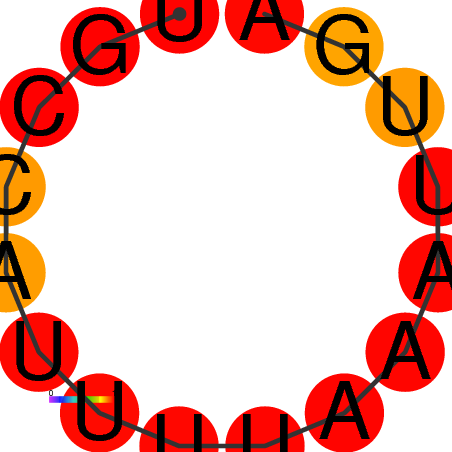 |
| dG=-0.9, p-value=0.009901 | dG=0.0, p-value=0.009901 |
|---|---|
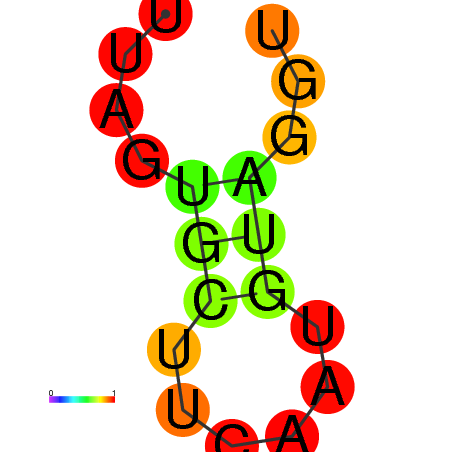 |
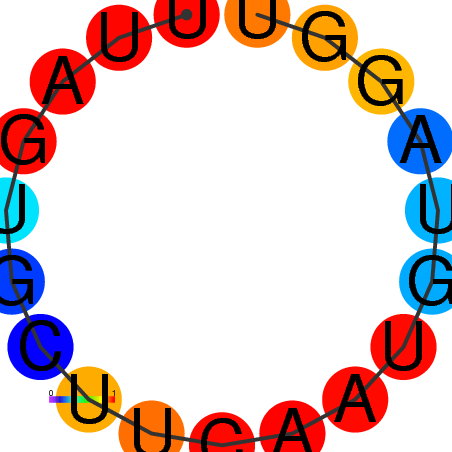 |
Generated: 03/07/2013 at 07:08 PM