

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:22511281-22511406 - | TTATATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGCGAGCATCTA----TCCAAA |
| droSim1 | chr3L:21812307-21812432 - | TTGTATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATCTA----TCCAAA |
| droSec1 | super_11:2446955-2447080 - | TTGTATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATCTA----TCCAAA |
| droYak2 | chr3L:22973852-22973977 - | TCATATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--ATATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCAACTA----TCCAAA |
| droEre2 | scaffold_4784:22180738-22180863 - | CCATATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATCTA----TCCAAA |
| droAna3 | scaffold_13337:22247678-22247807 - | TAAGATTCG---TAATATAGTAATATATACTGCCTACAATGGCCCTTAGTTTTCGACGTGTTTTGGTGTGCAGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATC------------ |
| dp4 | chrXR_group8:186470-186597 - | ACCCACCAG---TAA-----------ATACTGTCCACAATGGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT-GATATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GCGGCATGAGCATCTA----ACCAAA |
| droPer1 | super_80:80367-80497 + | ACAACCCACCAGAAT-----------ATACTGTCCACAATGGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT-GATATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GCGGCATGAGCATCTA----ACCAAA |
| droWil1 | scaffold_180698:7262478-7262610 + | TCTCTCGCA---TCATA-------------TATCCGCAATGGACCTTAGTTTTCGACGTGTTTTGGTGTGCTTAGGAAGTTTTCTATTCCGATTGAAACCGTCCAAAACTGAGGCCAATTGTTGTGATAGAAGCAACCATGAAAACAAA |
| droVir3 | scaffold_13049:17242400-17242523 + | TTATTTCCTAACTAG-----------ATAATGCCCACAATGGTTCTTAGTTTTAGACGTGTTTTGGTGTGCTTG-A-AGTT--TTGTTCCGATTGAAACCGTCCAAAACTGAGGCCGATTGT-GAGGTAAAAGCGTCCA---------A |
| droMoj3 | scaffold_6680:22273688-22273794 + | ------------------------------TGCCCACAATGGATCTTAGTTTTAGACGTGTTTTGGTGTGCTTGGG-AGTT--TTGTTCCGATTGAAACCGTCCAAAACTGAGGCCAATTGT-GAGGCGAAAGCATCCA----G----- |
| droGri2 | scaffold_15110:14942014-14942124 - | T------------------------------ACCCACAATGGATCTTAGTTTTAGACGTGTTTTGGTGTGCTTGGG-AGTT--TTGTTCCGATTGAAACCGTCCAAAACTGAGGCCAATTGT-GTGGTAAAAGCATCCA----AATATC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:22511281-22511406 - | TTATATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGCGAGCATCTA----TCCAAA |
| droSim1 | chr3L:21812307-21812432 - | TTGTATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATCTA----TCCAAA |
| droSec1 | super_11:2446955-2447080 - | TTGTATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATCTA----TCCAAA |
| droYak2 | chr3L:22973852-22973977 - | TCATATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--ATATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCAACTA----TCCAAA |
| droEre2 | scaffold_4784:22180738-22180863 - | CCATATCCG---TAGTGT------------TGTCCACAATAGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATCTA----TCCAAA |
| droAna3 | scaffold_13337:22247678-22247807 - | TAAGATTCG---TAATATAGTAATATATACTGCCTACAATGGCCCTTAGTTTTCGACGTGTTTTGGTGTGCAGGGG-AGTT--CTATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GAGGCGTGAGCATC------------ |
| dp4 | chrXR_group8:186470-186597 - | ACCCACCAG---TAA-----------ATACTGTCCACAATGGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT-GATATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GCGGCATGAGCATCTA----ACCAAA |
| droPer1 | super_80:80367-80497 + | ACAACCCACCAGAAT-----------ATACTGTCCACAATGGACCTTAGTTTTCGACGTGTTTTGGTGTGCTGGGG-AGTT-GATATTCCGATTGAAACCGTCCAAAACTGAGGCCAACTGT-GCGGCATGAGCATCTA----ACCAAA |
| droWil1 | scaffold_180698:7262478-7262610 + | TCTCTCGCA---TCATA-------------TATCCGCAATGGACCTTAGTTTTCGACGTGTTTTGGTGTGCTTAGGAAGTTTTCTATTCCGATTGAAACCGTCCAAAACTGAGGCCAATTGTTGTGATAGAAGCAACCATGAAAACAAA |
| droVir3 | scaffold_13049:17242400-17242523 + | TTATTTCCTAACTAG-----------ATAATGCCCACAATGGTTCTTAGTTTTAGACGTGTTTTGGTGTGCTTG-A-AGTT--TTGTTCCGATTGAAACCGTCCAAAACTGAGGCCGATTGT-GAGGTAAAAGCGTCCA---------A |
| droMoj3 | scaffold_6680:22273688-22273794 + | ------------------------------TGCCCACAATGGATCTTAGTTTTAGACGTGTTTTGGTGTGCTTGGG-AGTT--TTGTTCCGATTGAAACCGTCCAAAACTGAGGCCAATTGT-GAGGCGAAAGCATCCA----G----- |
| droGri2 | scaffold_15110:14942014-14942124 - | T------------------------------ACCCACAATGGATCTTAGTTTTAGACGTGTTTTGGTGTGCTTGGG-AGTT--TTGTTCCGATTGAAACCGTCCAAAACTGAGGCCAATTGT-GTGGTAAAAGCATCCA----AATATC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
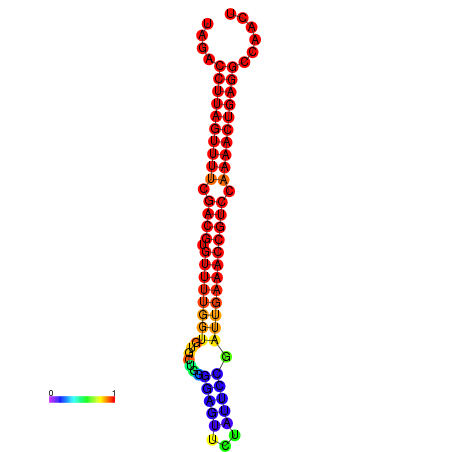 |
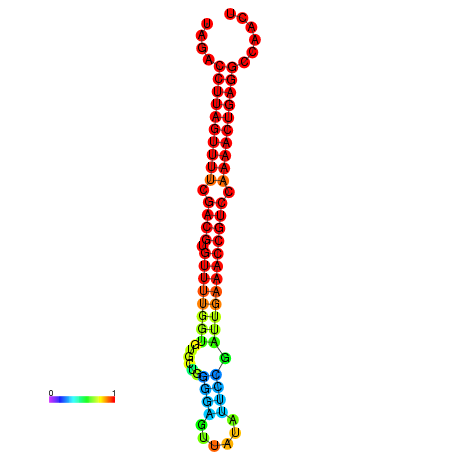
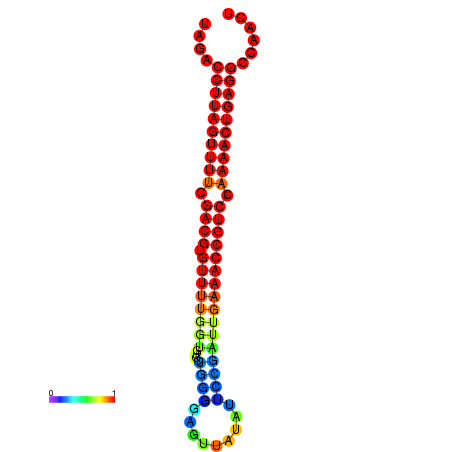
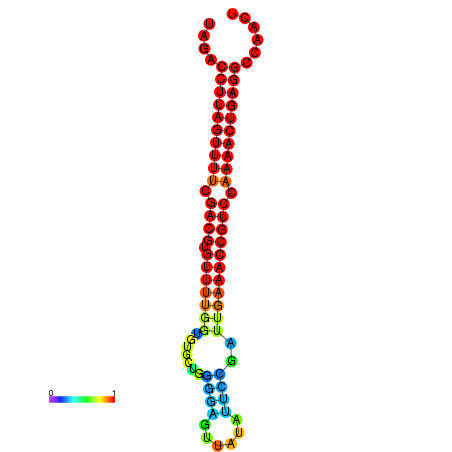
| dG=-24.5, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
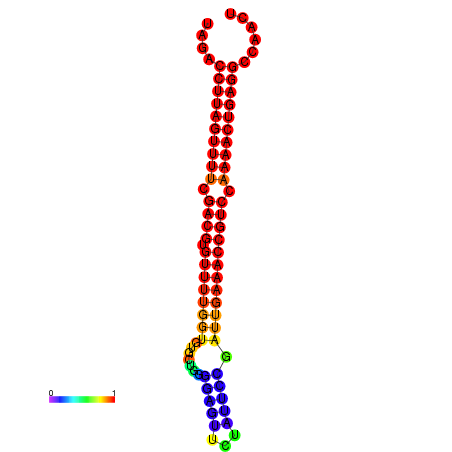 |
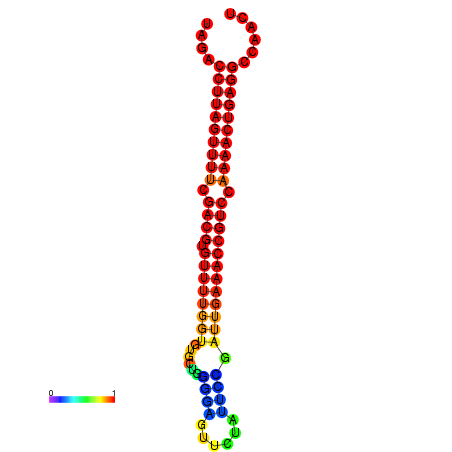
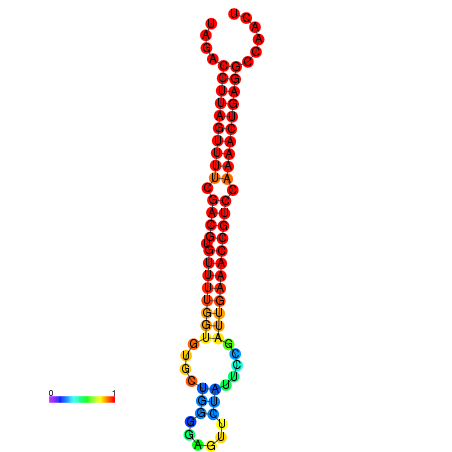
| dG=-28.2, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|---|---|---|
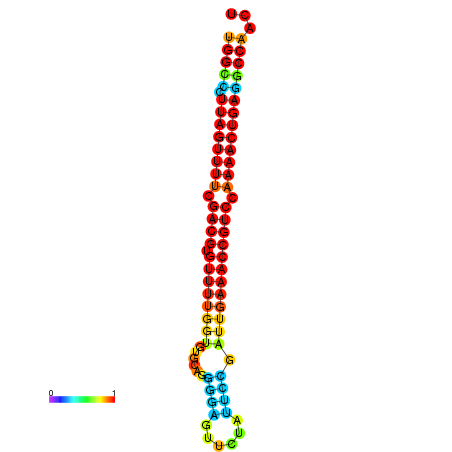 |
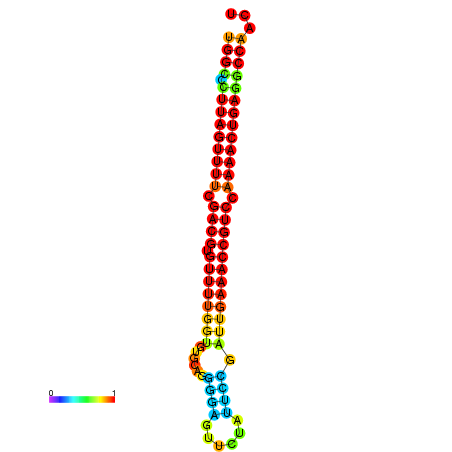 |
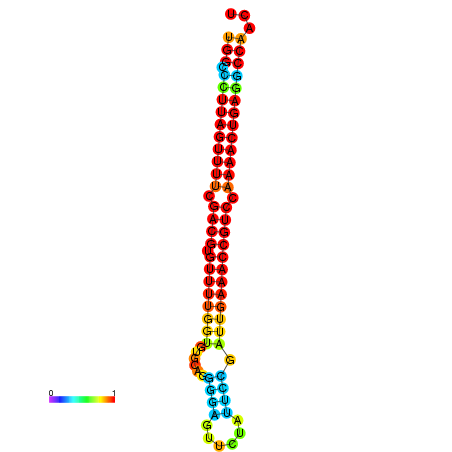 |
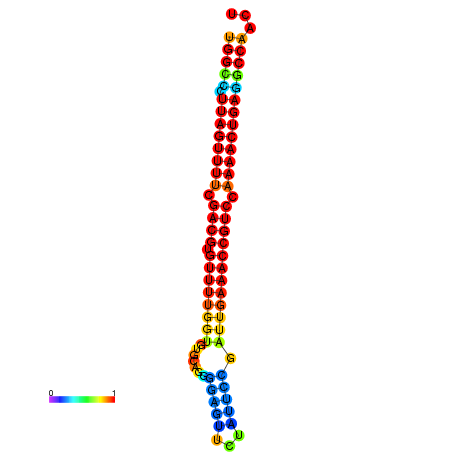 |
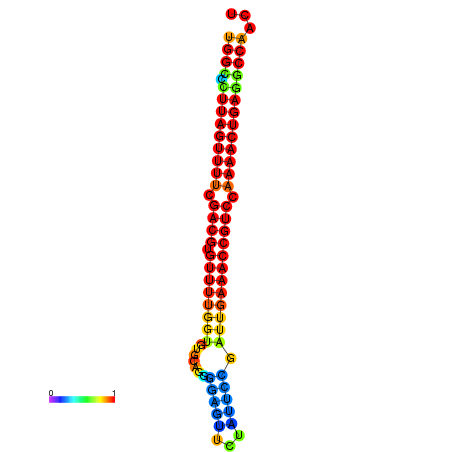 |
| dG=-29.1, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.5, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-29.1, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.5, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-30.9, p-value=0.009901 |
|---|
 |
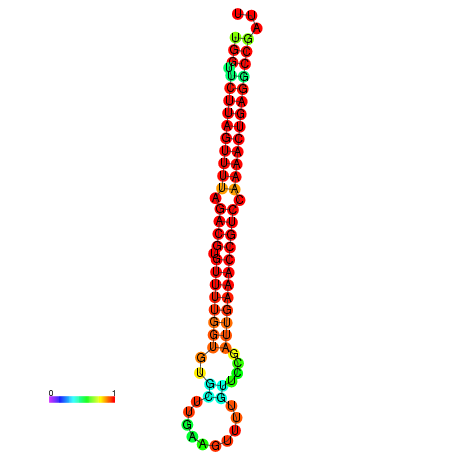
| dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 |
|---|---|---|---|---|
 |
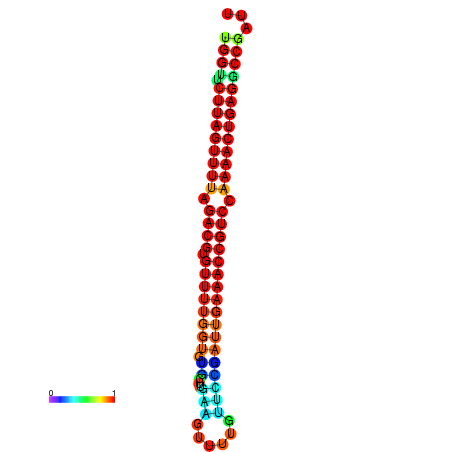
 |
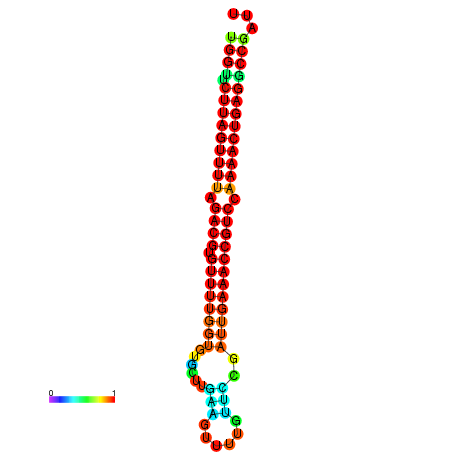
 |
 |
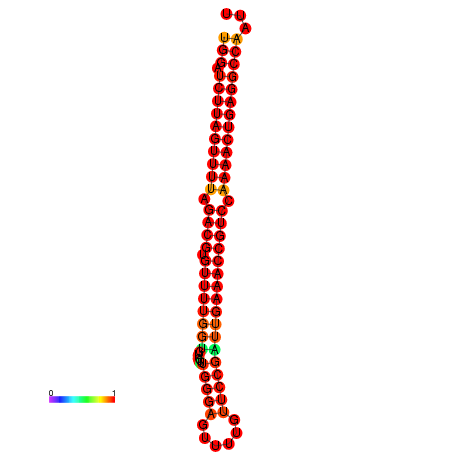
 |
| dG=-27.6, p-value=0.009901 | dG=-27.5, p-value=0.009901 |
|---|---|
 |
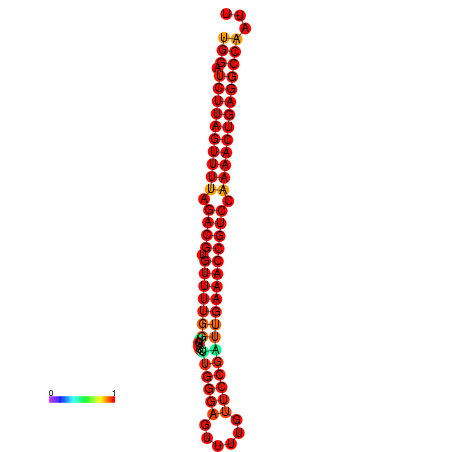 |
| dG=-27.6, p-value=0.009901 | dG=-27.5, p-value=0.009901 |
|---|---|
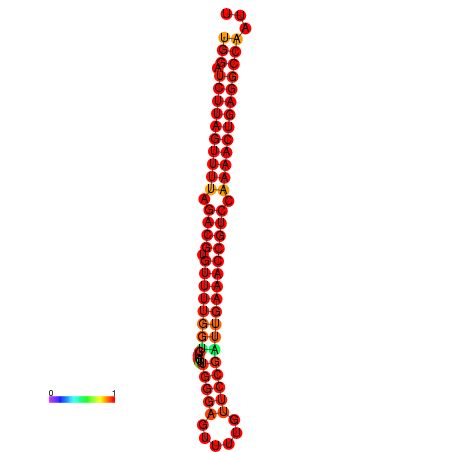 |
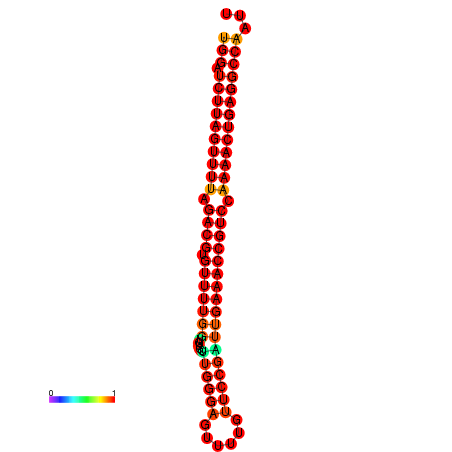 |
Generated: 03/07/2013 at 07:06 PM