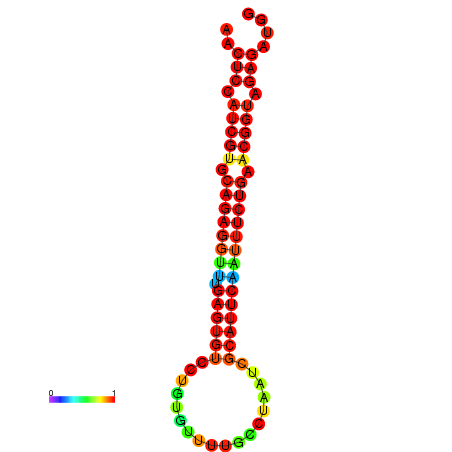
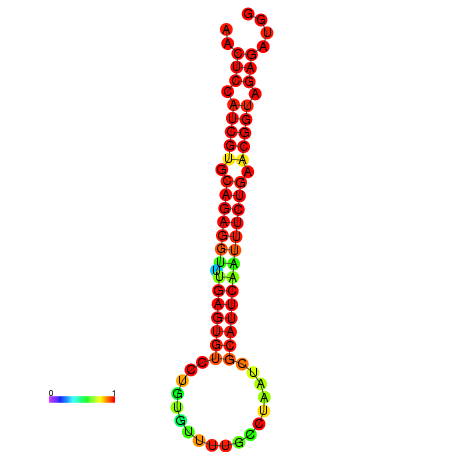
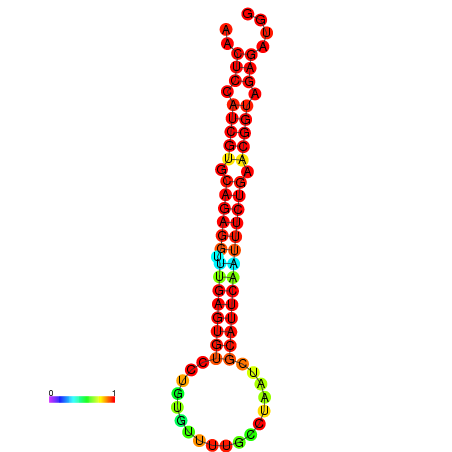
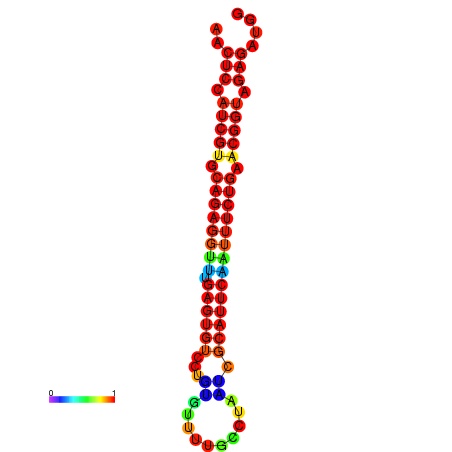
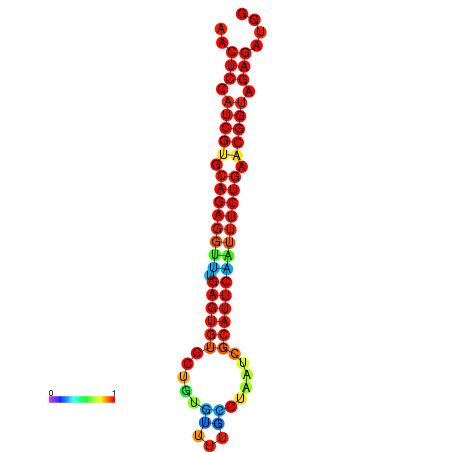
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:3299040-3299166 + | TAAGGAACACTTTTTGGCC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCCTGT---GTTTTGCCTAATCGCATTCAATTTCTGAACGGTAGAGATGGTACGCTTAGAAACAACCTAAA--ACAA-------------------------------------------------TT |
| droSim1 | chr3L:2834561-2834687 + | TAAAGAACACTTTTTGGCC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCCTGT---GTTTTGCCTAATCGCATTCAATTTCTGCACGGTAGAGATGGTACGCTTAGAAACAACCTAAA--AAAA-------------------------------------------------TT |
| droSec1 | super_2:3291343-3291466 + | TAATAAACACTTTTTGGTC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCCTGT---GTTTTGCCTAATCGCATTCAATTTCTGCAG---AGAGATGGTACGCTTAGAAACAACCTACA--AAAA-------------------------------------------------TT |
| droYak2 | chr3L:3844747-3844864 + | TAAAGTAAA---------C--AGCTAATCAACTCCATCGTGCAGAAGTTTGAGTGTCCTGT---GTGTTGCCTAATCGCATTCAATTTCTGAACGGTAGAGATGGTACGCTTAGAAACAACCTAAA--ATGA-------------------------------------------------GT |
| droEre2 | scaffold_4784:5981989-5982116 + | AAGGAATACACTTTTGTCC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCTTGT---GTGTTGCCTAATCGCATTCAATTTCTGAACGGTAGAGATGGTACGCTTAGAAACAACCTAAA--GCAA-------------------------------------------------GT |
| droAna3 | scaffold_13337:7606422-7606539 + | TGAAGGATGTTTTTTGAGA--AGCTCATCAGCTCCATCGTGCAGAGGTTTGAGTGTCTAAT----TTTTAACCATTCGCATTCAACCTTTGGACGGTAGAGATGGTCAGTGAGAAAGCAACCTA----------------------------------------------------------- |
| dp4 | chr4_group1:2379981-2380081 + | AATT----------TAAAA--AAATGAATAAATTCAAATGACAAAAATGTAAATGAATTAA-----------AAAATAAATTCAAATGATAAAAAGTAAA----------TGAATAAAAAAATAAAGTAAAA-------------------------------------------------TT |
| droPer1 | Unknown | --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 | scaffold_180698:10215726-10215851 + | ATTCAAATACTTTTGCGTTAAGGTTGGCTGGCTCCATCGTGCTTAGGTTTGAGTGTCTTCGTTTGTTTTCTCTAATGGCATTCAACCTTGGAGCGATAGAGACGGCCAATCGAGAAACAGCCTAAA--------------------------------------------------------- |
| droVir3 | scaffold_12758:855562-855687 - | TATGTATGGATCTCTCATC--AGTTAGCTAGCTCCATCGTGCACAGGTTTGAGTGTT-TGA---GTTTTTCCTAGTGGCATTCAACCTTTGAACGGTAGAGATGGCCAACTGAAAAGCGACCTAAA--CTAA-------------------------------------------------TA |
| droMoj3 | scaffold_6680:1909563-1909732 - | TCAGGTATCCAATATG-----GGTTAGCGAGCTCCATCGTGCACAGGTTTGAGTGTT-TGT---GT-TTGGCTAGTCGCATTCAGCCTTTGAACGGTAGAGATGGCCAACTGAAAAGCGACCTAAC---TGAAAGCGAATCAAGGGAACTAGGCCAATAGGCGAGAATCATATAGAAATAGTT |
| droGri2 | scaffold_15110:657067-657177 + | TAAAG-----------------GTTGGCGAACTCCATCGTGCACGGGTTTGAGTGTC-TGA---GT-TTGCCAAGTGGCATTCAGCCCCTGGACGCTAGAGATGGCCAACTGAAAAGCAACCTAAA--AA------------------------------------------------CAATC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:3299040-3299166 + | TAAGGAACACTTTTTGGCC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCCTGT----GTTTTGCCTAATCGCATTCAATTTCTGAACGGTAGAGATGGTACGCTTAGAAACAACCTAAA------------------------------------------------ACAATT |
| droSim1 | chr3L:2834561-2834687 + | TAAAGAACACTTTTTGGCC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCCTGT----GTTTTGCCTAATCGCATTCAATTTCTGCACGGTAGAGATGGTACGCTTAGAAACAACCTAAA------------------------------------------------AAAATT |
| droSec1 | super_2:3291343-3291466 + | TAATAAACACTTTTTGGTC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCCTGT----GTTTTGCCTAATCGCATTCAATTTCTGCAG---AGAGATGGTACGCTTAGAAACAACCTACA------------------------------------------------AAAATT |
| droYak2 | chr3L:3844747-3844864 + | TAAAGTAAA---------C--AGCTAATCAACTCCATCGTGCAGAAGTTTGAGTGTCCTGT----GTGTTGCCTAATCGCATTCAATTTCTGAACGGTAGAGATGGTACGCTTAGAAACAACCTAAA------------------------------------------------ATGAGT |
| droEre2 | scaffold_4784:5981989-5982116 + | AAGGAATACACTTTTGTCC--AGCTAATCAACTCCATCGTGCAGAGGTTTGAGTGTCTTGT----GTGTTGCCTAATCGCATTCAATTTCTGAACGGTAGAGATGGTACGCTTAGAAACAACCTAAA------------------------------------------------GCAAGT |
| droAna3 | scaffold_13337:7606422-7606539 + | TGAAGGATGTTTTTTGAGA--AGCTCATCAGCTCCATCGTGCAGAGGTTTGAGTGTCTAAT-----TTTTAACCATTCGCATTCAACCTTTGGACGGTAGAGATGGTCAGTGAGAAAGCAACCTA-------------------------------------------------------- |
| droWil1 | scaffold_180698:10215726-10215851 + | ATTCAAATACTTTTGCGTTAAGGTTGGCTGGCTCCATCGTGCTTAGGTTTGAGTGTCTT-CGTTTGTTTTCTCTAATGGCATTCAACCTTGGAGCGATAGAGACGGCCAATCGAGAAACAGCCTAAA------------------------------------------------------ |
| droVir3 | scaffold_12758:855562-855687 - | TATGTATGGATCTCTCATC--AGTTAGCTAGCTCCATCGTGCACAGGTTTGAGTGTT-TGA----GTTTTTCCTAGTGGCATTCAACCTTTGAACGGTAGAGATGGCCAACTGAAAAGCGACCTAAA------------------------------------------------CTAATA |
| droMoj3 | scaffold_6680:1909563-1909732 - | TCAGGTATCCAATATG-----GGTTAGCGAGCTCCATCGTGCACAGGTTTGAGTGTT-TGT----GT-TTGGCTAGTCGCATTCAGCCTTTGAACGGTAGAGATGGCCAACTGAAAAGCGACCTAACTGAAAGCGAATCAAGGGAACTAGGCCAATAGGCGAGAATCATATAGAAATAGTT |
| droGri2 | scaffold_15110:657067-657177 + | TAAAG-----------------GTTGGCGAACTCCATCGTGCACGGGTTTGAGTGTC-TGA----GT-TTGCCAAGTGGCATTCAGCCCCTGGACGCTAGAGATGGCCAACTGAAAAGCAACCTAAA-----------------------------------------------AACAATC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-21.1, p-value=0.009901 |
|---|---|---|---|---|
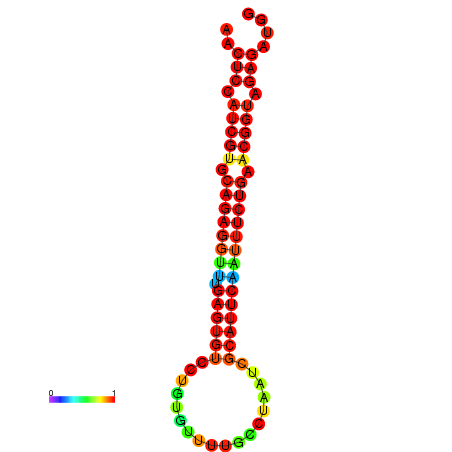 |
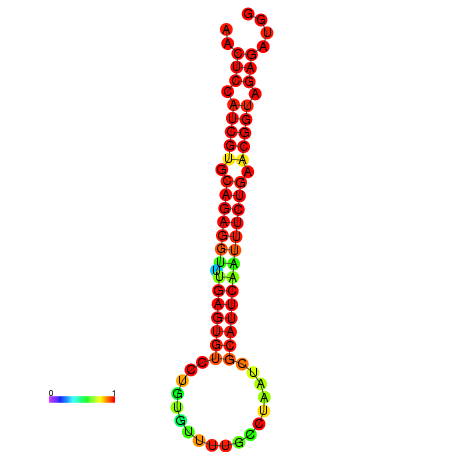 |
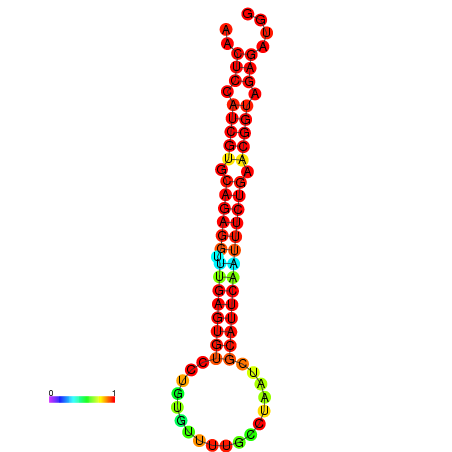 |
 |
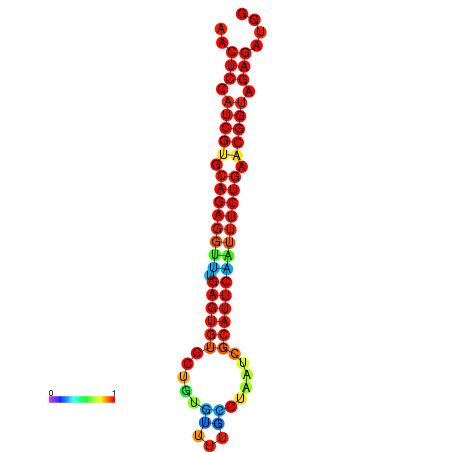 |
| dG=-28.1, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-28.1, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|---|---|---|
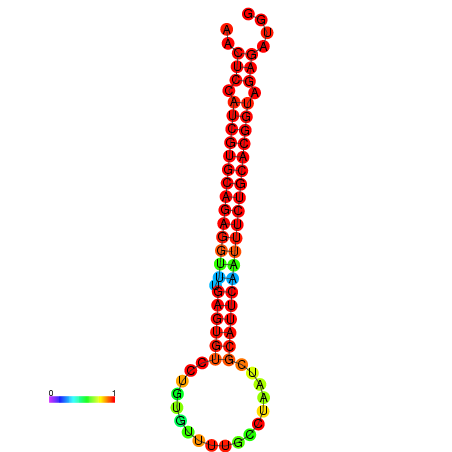 |
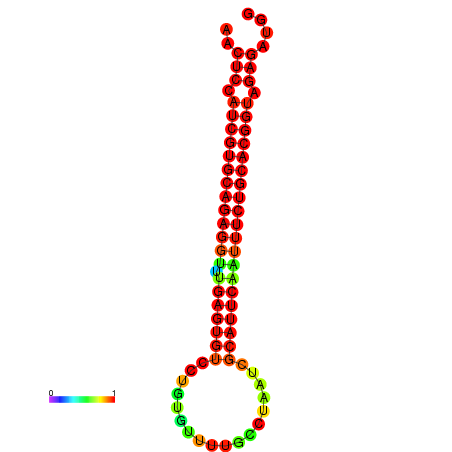 |
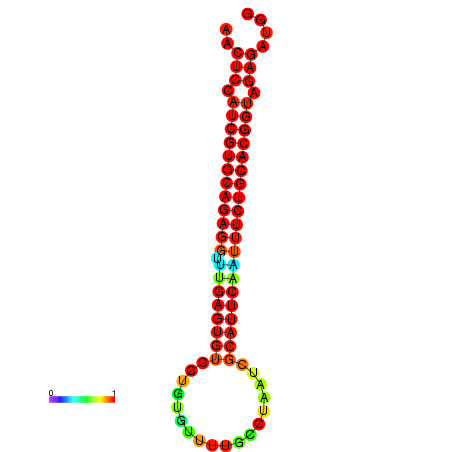 |
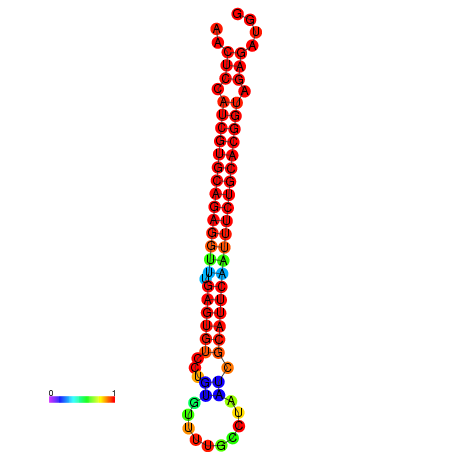 |
 |
| dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.9, p-value=0.009901 |
|---|---|---|---|---|
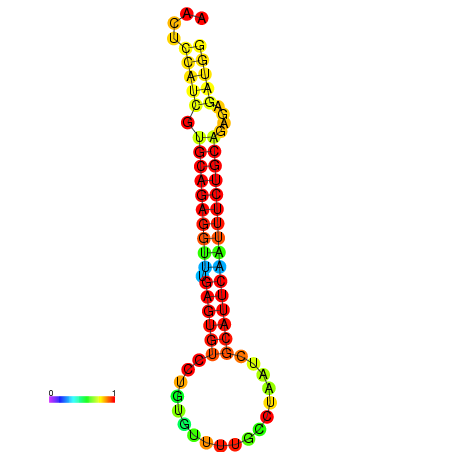 |
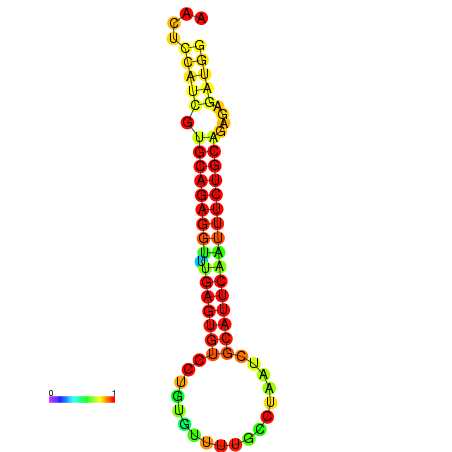 |
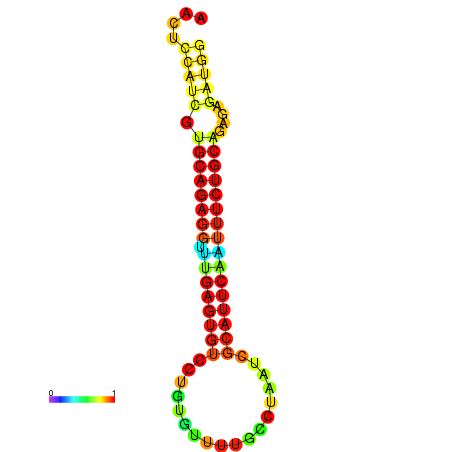 |
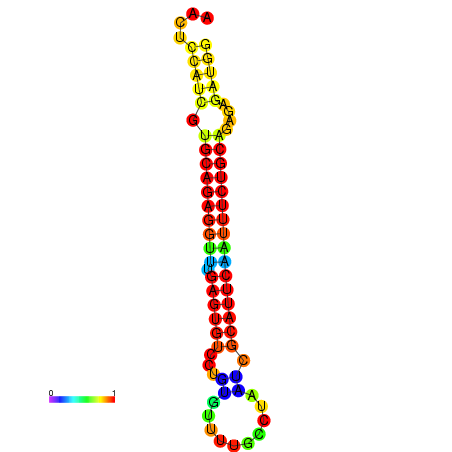 |
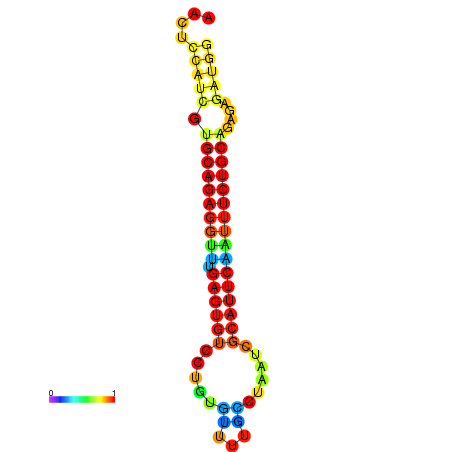 |
| dG=-21.8, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 |
|---|---|---|---|---|
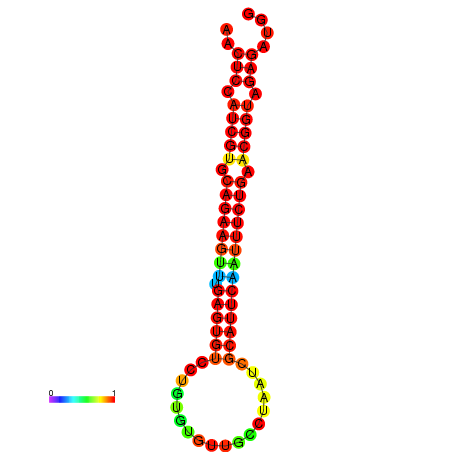 |
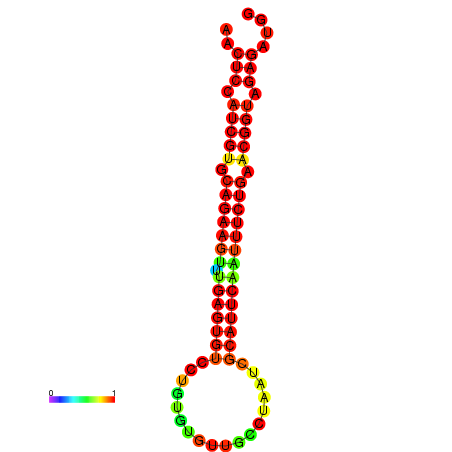 |
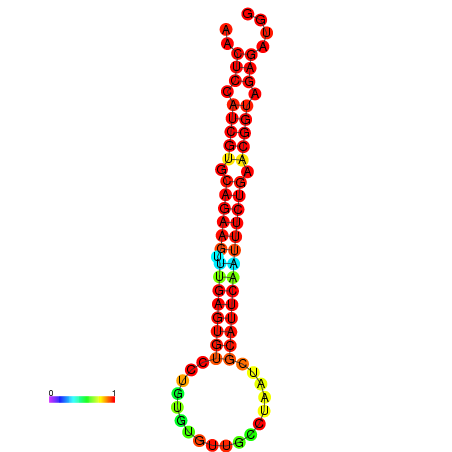 |
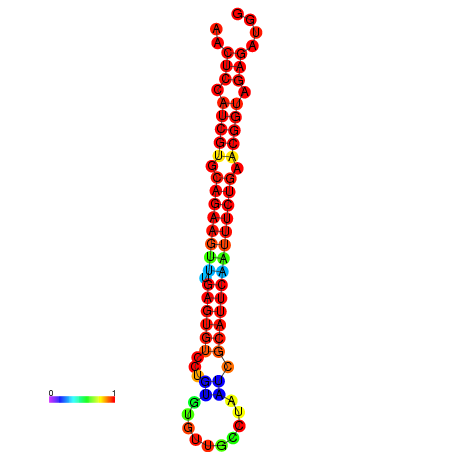 |
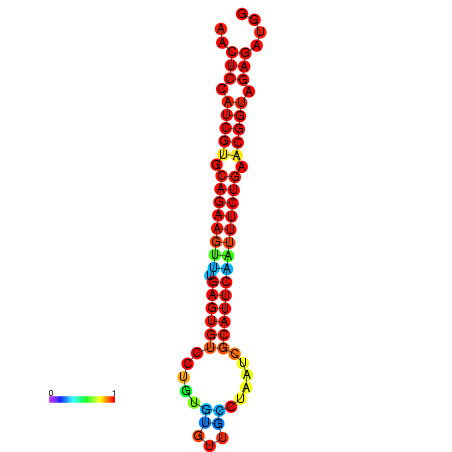 |
| dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 |
|---|---|---|---|---|
 |
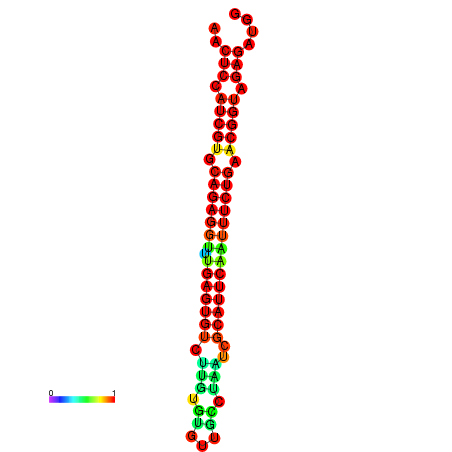 |
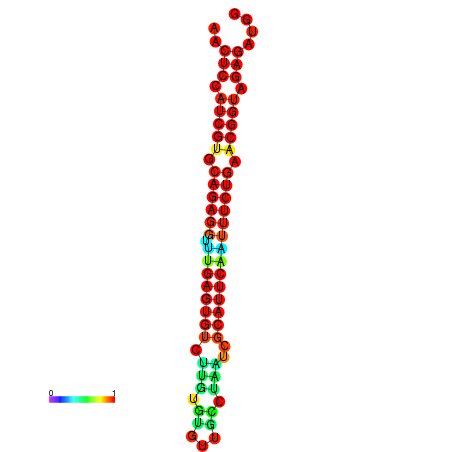 |
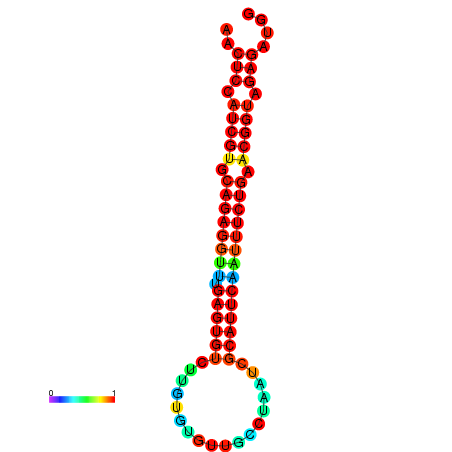 |
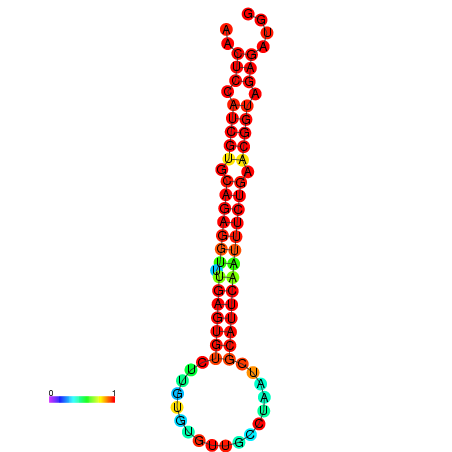 |
| dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 |
|---|---|---|---|---|
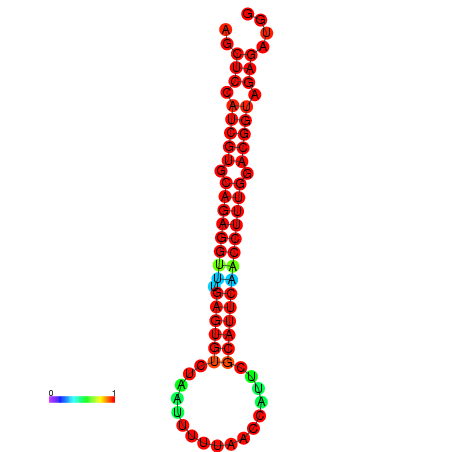 |
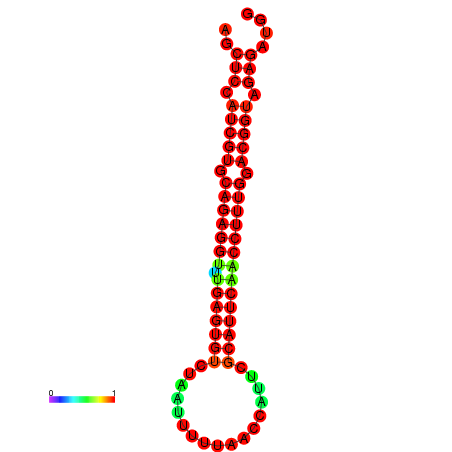 |
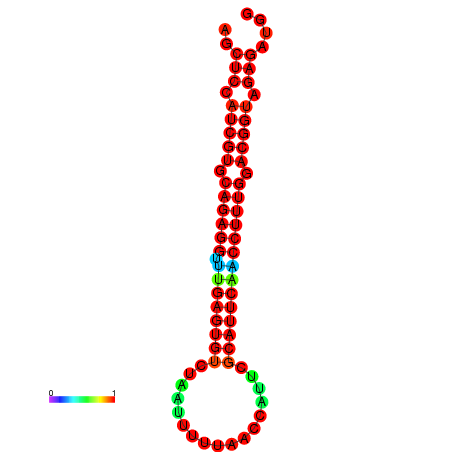 |
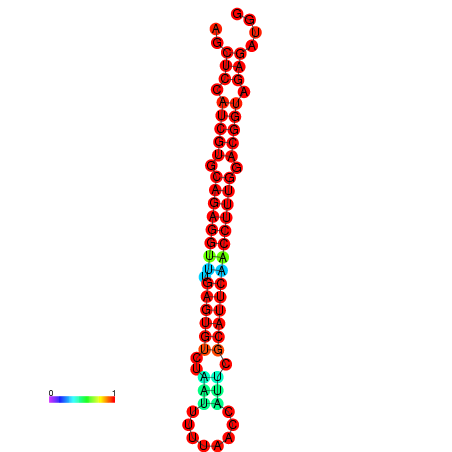 |
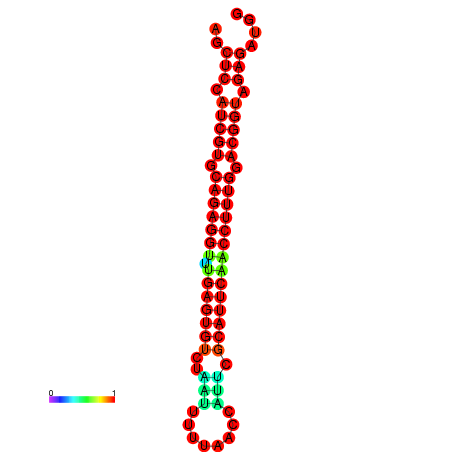 |
| dG=-3.4, p-value=0.009901 | dG=-2.9, p-value=0.009901 | dG=-2.7, p-value=0.009901 | dG=-2.7, p-value=0.009901 |
|---|---|---|---|
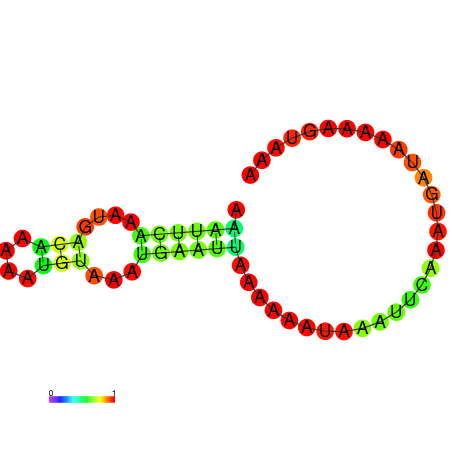 |
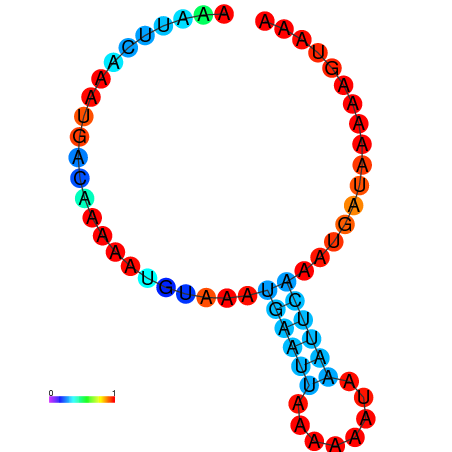 |
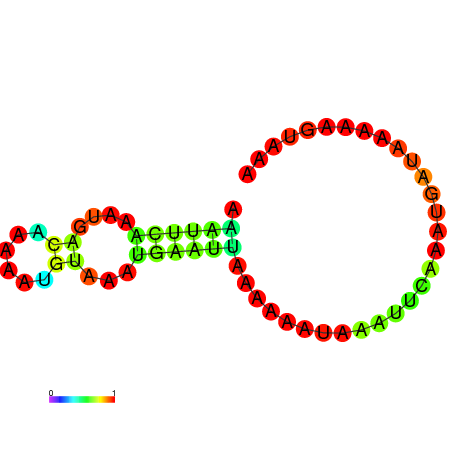 |
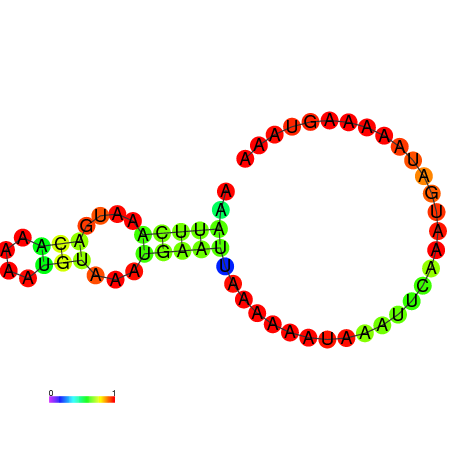 |
No secondary structure predictions returned
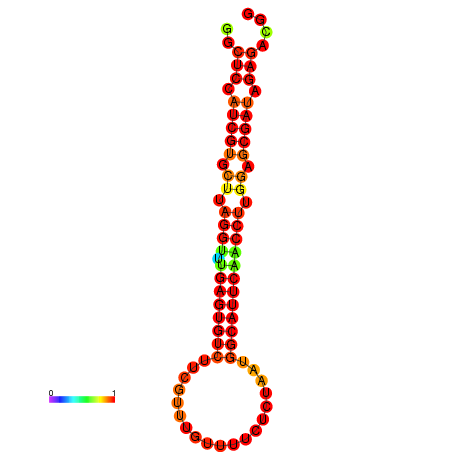
| dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 | dG=-24.5, p-value=0.009901 |
|---|---|---|
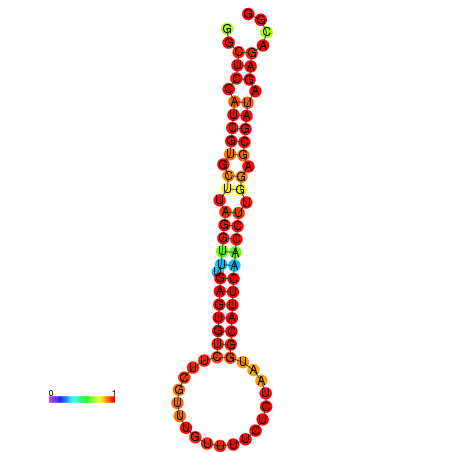 |
 |
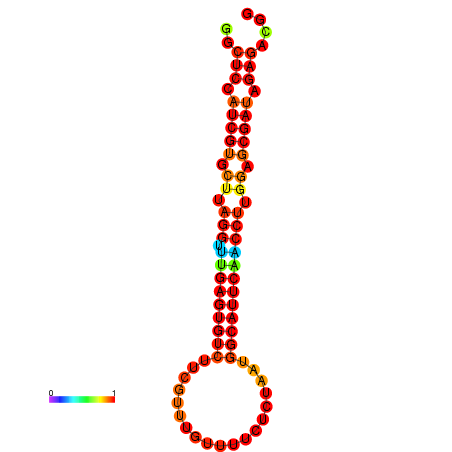 |
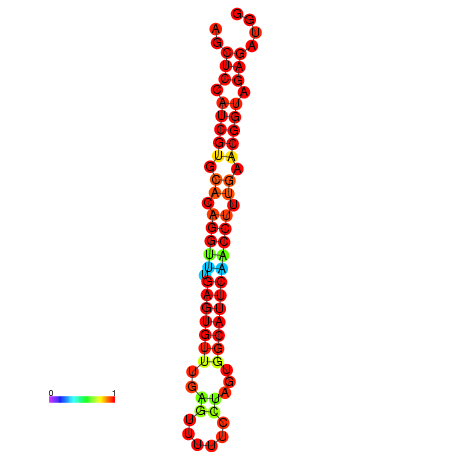
| dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
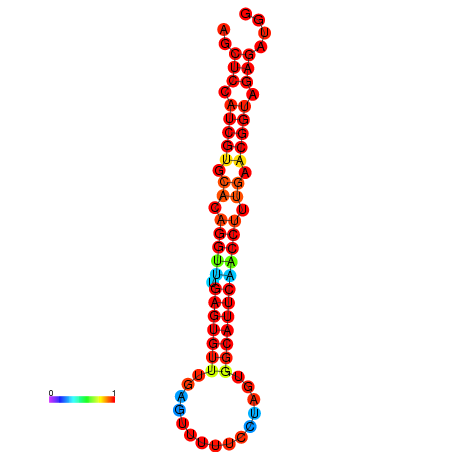 |
 |
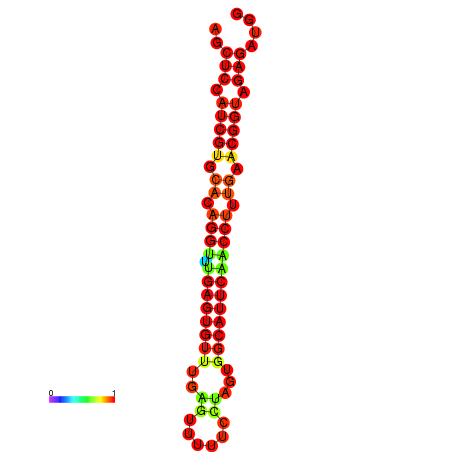
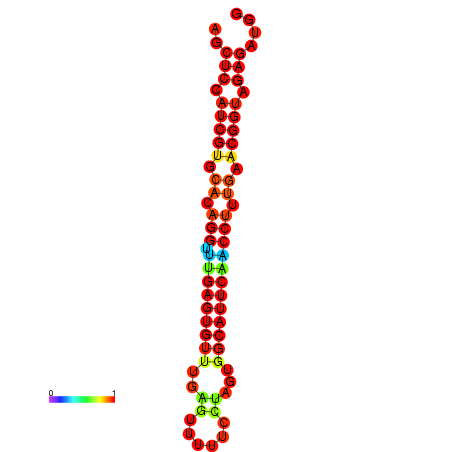
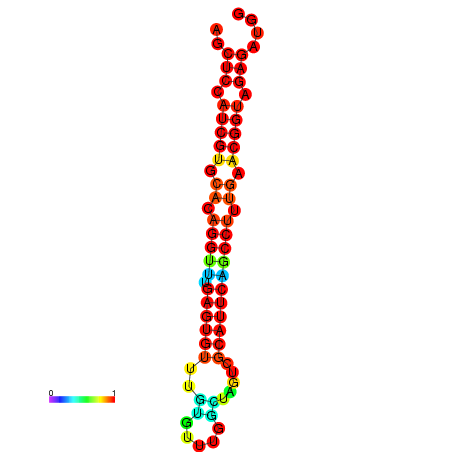
| dG=-20.9, p-value=0.009901 | dG=-20.9, p-value=0.009901 | dG=-20.9, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.5, p-value=0.009901 |
|---|---|---|---|---|
 |
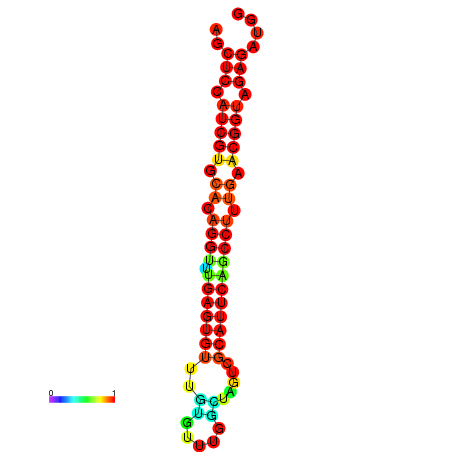 |
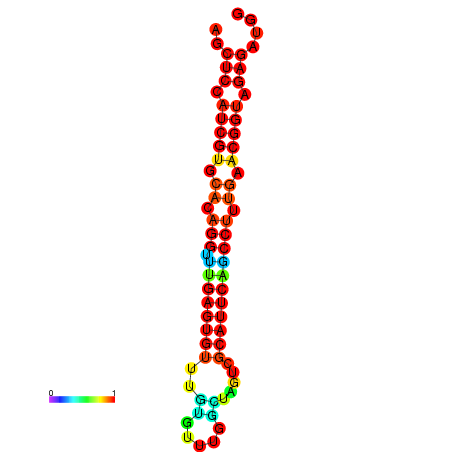 |
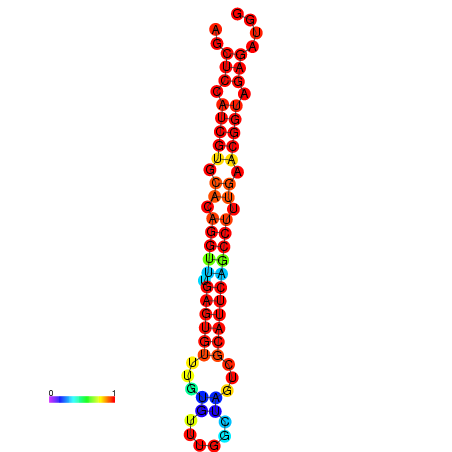 |
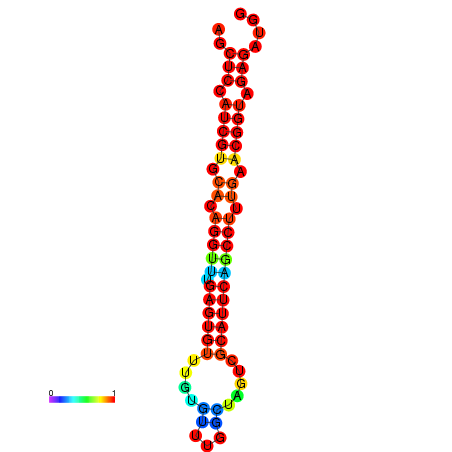 |
| dG=-24.6, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.6, p-value=0.009901 | dG=-24.2, p-value=0.009901 |
|---|---|---|---|
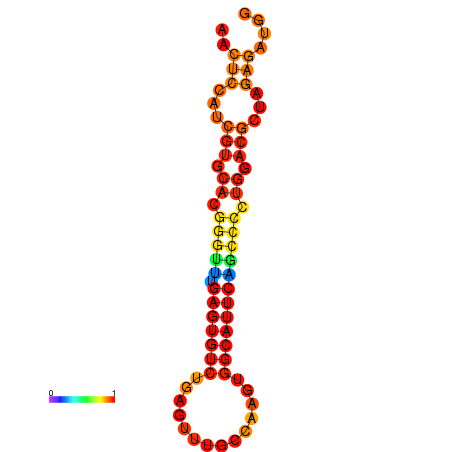 |
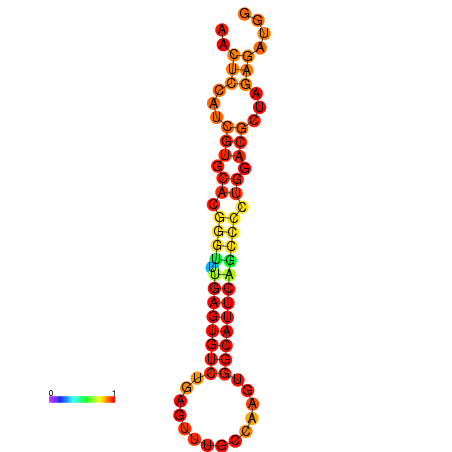 |
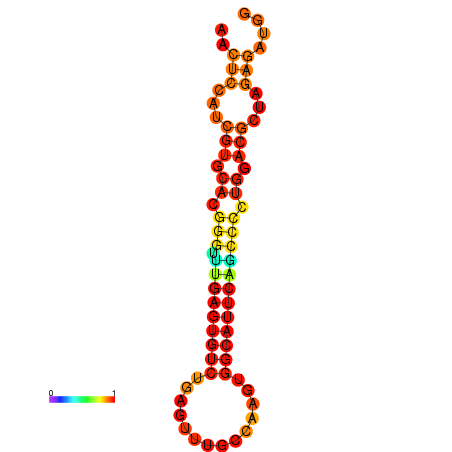 |
 |
Generated: 03/07/2013 at 07:05 PM