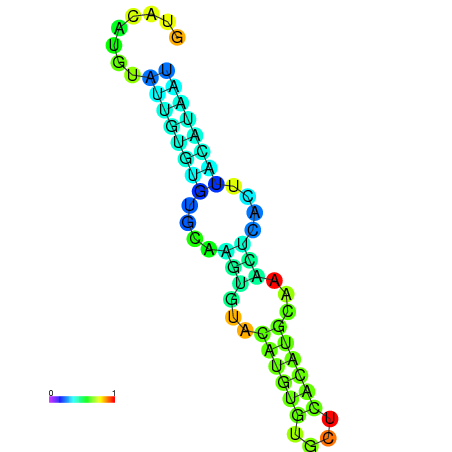
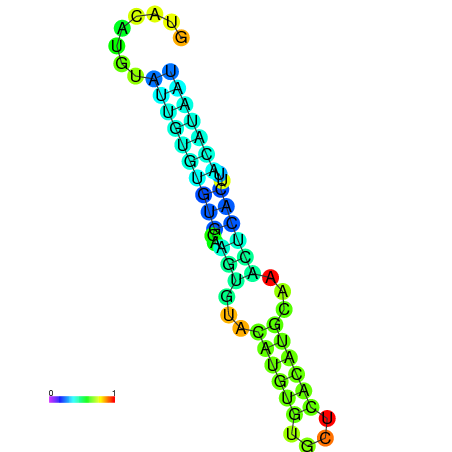
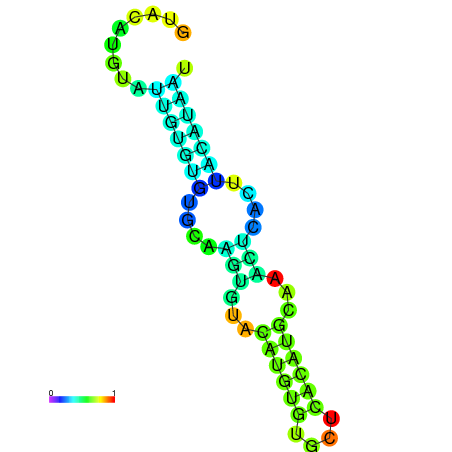
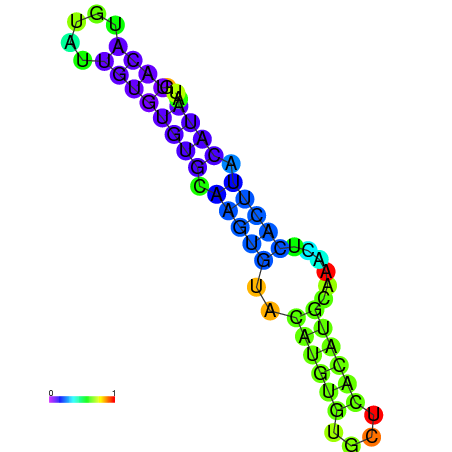
| dm3 |
chr4:646737-646853 + |
GACTCATGCA--------------------CGTCT-TACAAACACAAGATTTTCTGGGTGTTGCGTTGTG------------TG--------TACCTGTGTACAGGCGTATTCACATGCA--------ACATCCCTTACATCTTGCTTGATTATACTCCGCCCG---------------------------------------------------------------CC |
| droSim1 |
chrU:391095-391211 + |
GACGCATGCA--------------------CGTCT-TACAAACACAAGAATTTCTGCGCGTAGAGTTTTG------------TG--------TACATGTGTACGTGCGTATTCACATCCA--------ACATCCCTGACGTCTTGCTTGATTATACTCCGCCCG---------------------------------------------------------------CC |
| droSec1 |
super_30:369242-369358 + |
GACGCATGCA--------------------CGTCT-TACAAACACAAGAATTTCTGCGCGTAGAGTTTTG------------TG--------TACATGTGTACGTGCGTATTTACATCCA--------ACATCCCTGACGTCTTGCTTGATTATACTCCGCCCG---------------------------------------------------------------CC |
| droYak2 |
chr4:700584-700681 + |
GACTCATGCA--------------------CGTCTTTACAAACACAAGATATTCTACGCGTTGTGTTTTG------------TG--------TACAT--------------------GCA--------ACATCCCTGACGTCTTGCGCGATTATACTCTGCCCG---------------------------------------------------------------TC |
| droEre2 |
scaffold_4512:633126-633238 - |
GAGACATGCA--------------------CGTCTTTACAAACACAAGATATTCTGCGCGTTGTGTTTTG-------------ACATGTATGTACAT--------------------GCAACCGTTATGCGTTCCAAGGACCCTATTAGATCGGAACACGGTCA---------------------------------------------------------------GT |
| droAna3 |
scaffold_13337:22987804-22987813 + |
TCCGCCCGC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T |
| dp4 |
chrXR_group8:8504771-8504835 + |
CT----------------------------------TACAAATTCTAGAATTTCTGTCTGTCTGTCTGTCTGTTTTTGTGTGTG--------TATGT----GTGTGTGTGC---------------------------------------------------------------------------------------------------------------------- |
| droPer1 |
super_150:7787-7872 - |
AATGCATACA--------------------CGATA-TACATACATATGTGTGTGTGTGTGTGTGTGTGTG------------TG--------TGTGTGTGTGTGTGTG----------TA--------GACGTATGTGTAT-----GTGA------------------------------------------------------------------------------- |
| droWil1 |
scaffold_181130:1758155-1758267 + |
CACTTTTGTG--------------------CTGCG-TGTGAGT----GTGTGTGTGTGAGTTTAAGTGTG------------TG--------TATGTGCGAACATACGAATTTAGATAAA--------GATTCGTATACACATGACATTCGCATACTCACACCT---------------------------------------------------------------AC |
| droVir3 |
scaffold_13049:11239836-11239930 - |
TACATACGCA--------------------CA----TACATACATAGGTGTTCGTATG----------TA------------TG--------TACGTATGTAGGTGTG--CT---ATACACCCGTCAAAAGAGCTAATTTCCTTTCTTGGTTAT--------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6500:15364731-15364841 + |
GGGTCAGGCAAATTCGAATGTGAATTCAAAATACA-TACATACATATGTACAT---------GTATTGTG------------TG--------TGCAAGTGTACATGTGTGCTCACATGCA--------AACTCACTTACATAATGTATG-------------------------------------------------------------------------------- |
| droGri2 |
scaffold_14822:646607-646724 + |
AACAAATGCA--------------------GCGCA-TGT----------------TT------------------------------------------TGATTCACAAGCCTGAACATA--------TCCTAGCATAT------------C------------GTTGGTAACGGGGCTCTGCAGAATACACACACAAACATATTGATGCTCGCGTTGATTATCATGCC |