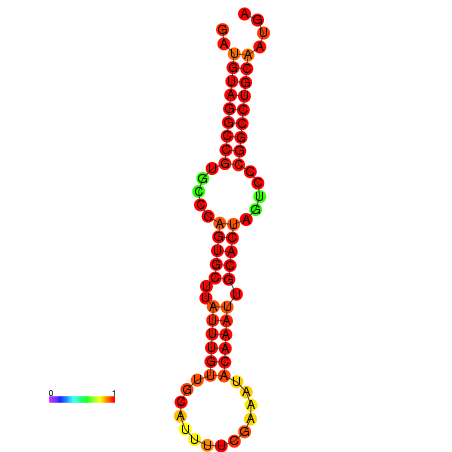
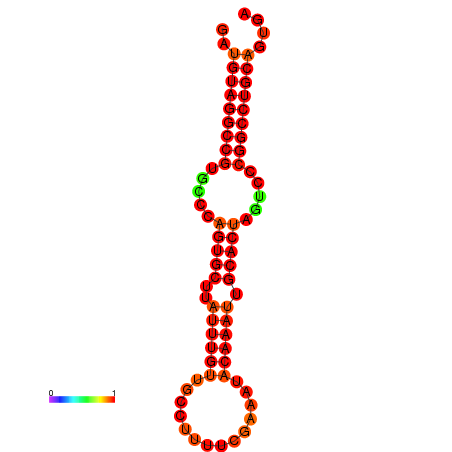
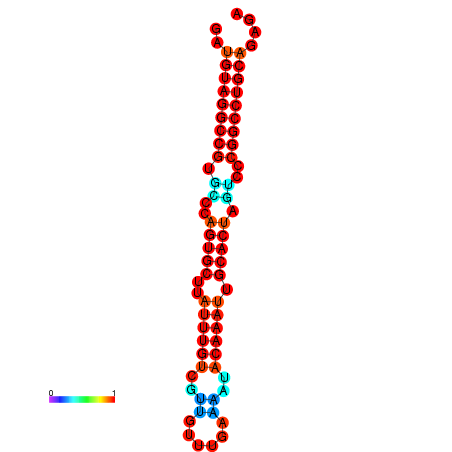
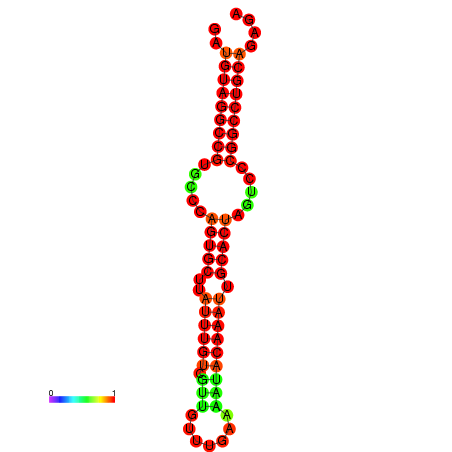
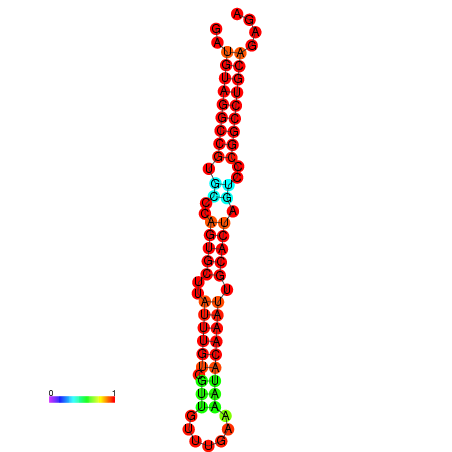
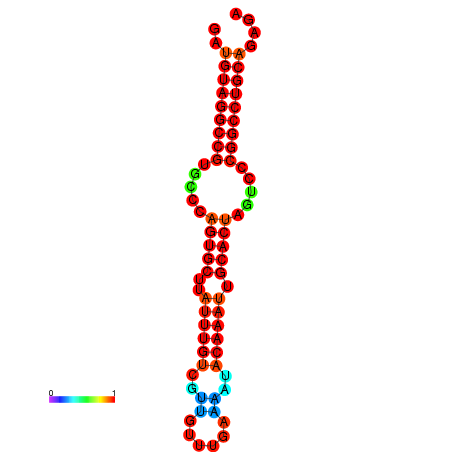
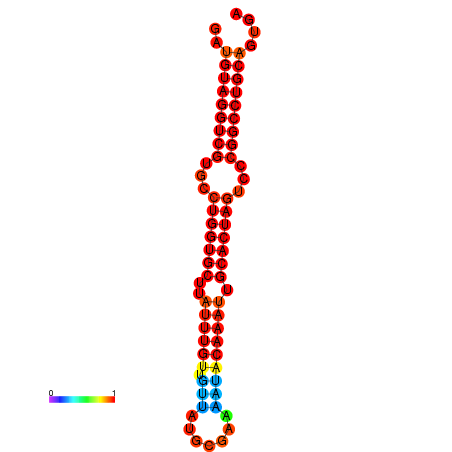
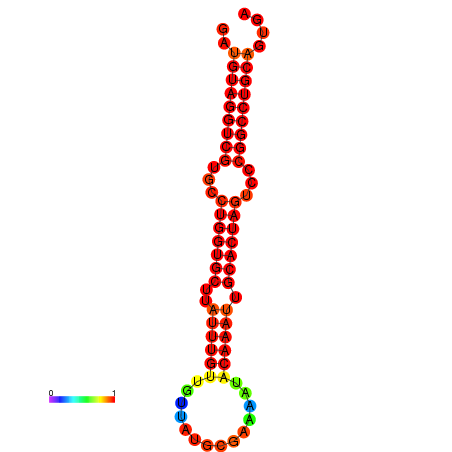
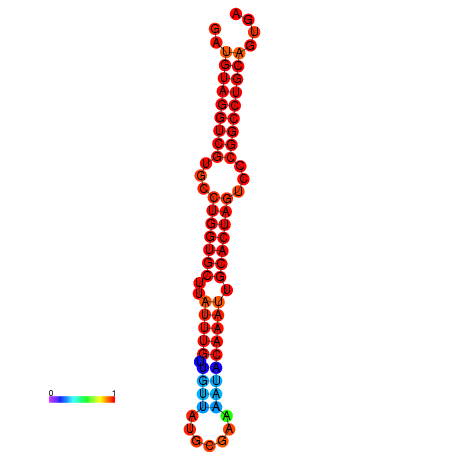
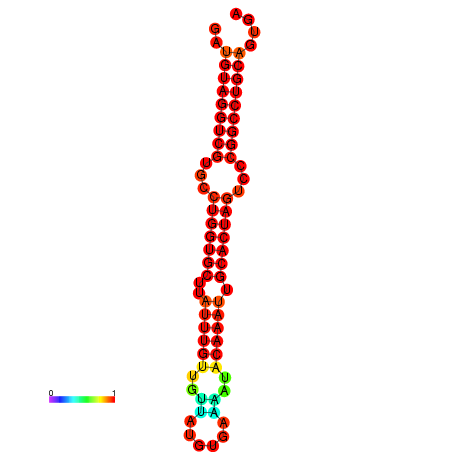
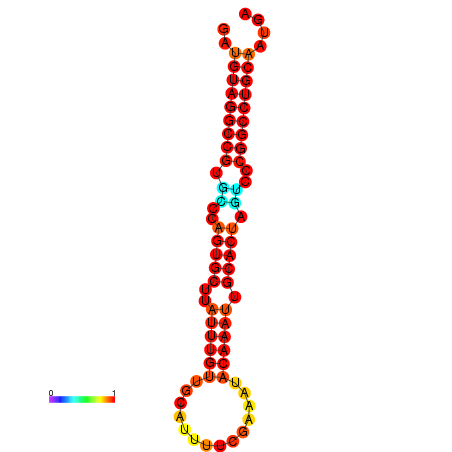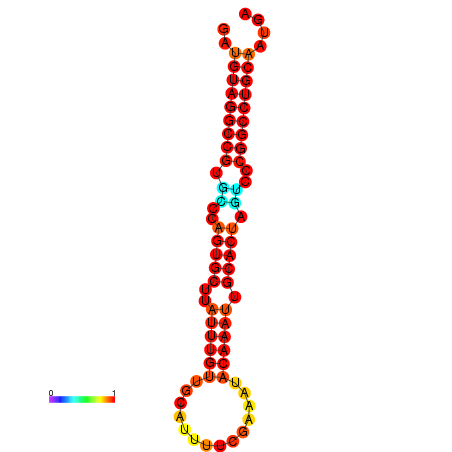| AGTGCAGT-GGTACATTAAAACGTCACCTGATGTAGGCCGTGCCCAGTGCTTATTTGTTGCCTTTTCGAAATACAAATTGCACTAGTCCCGGCCTGCAGTGAGTGTCGCAGTCGACGATCTACACAGTGCA | Size | Hit Count | Total Norm | Total | GSM343915
Embryo |
| ...........................................................................AATTGCACTAGTCCCGGCCTGC.................................. | 22 | 1 | 2817.00 | 2817 | 2817 |
| ...........................................................................AATTGCACTAGTCCCGGCCTG................................... | 21 | 1 | 2235.00 | 2235 | 2235 |
| ...........................................................................AATTGCACTAGTCCCGGCCT.................................... | 20 | 1 | 266.00 | 266 | 266 |
| ..................................AGGCCGTGCCCAGTGCTTATTTG.......................................................................... | 23 | 1 | 108.00 | 108 | 108 |
| ...........................................................................AATTGCACTAGTCCCGGCCTGCA................................. | 23 | 1 | 99.00 | 99 | 99 |
| ............................................................................ATTGCACTAGTCCCGGCCTGC.................................. | 21 | 1 | 16.00 | 16 | 16 |
| ..........................................................................AAATTGCACTAGTCCCGGCCTGC.................................. | 23 | 1 | 12.00 | 12 | 12 |
| ...........................................................................AATTGCACTAGTCCCGGCC..................................... | 19 | 1 | 12.00 | 12 | 12 |
| ............................................................................ATTGCACTAGTCCCGGCCTG................................... | 20 | 1 | 12.00 | 12 | 12 |
| ..........................................................................AAATTGCACTAGTCCCGGCCTG................................... | 22 | 1 | 7.00 | 7 | 7 |
| ..........................................................................AAATTGCACTAGTCCCGGCCT.................................... | 21 | 1 | 5.00 | 5 | 5 |
| ...........................................................................AATTGCACTAGTCCCGGC...................................... | 18 | 1 | 5.00 | 5 | 5 |
| ..................................AGGCCGTGCCCAGTGCTTATTTGT......................................................................... | 24 | 1 | 4.00 | 4 | 4 |
| ............................................................................ATTGCACTAGTCCCGGCCTGCA................................. | 22 | 1 | 2.00 | 2 | 2 |
| ..................................AGGCCGTGCCCAGTGCTTATTT........................................................................... | 22 | 1 | 2.00 | 2 | 2 |
| .........................................................TTGCCTTTTCGAAATACAA....................................................... | 19 | 1 | 1.00 | 1 | 1 |
| .........................................................TTGCCTTTTCGAAATACAAA...................................................... | 20 | 1 | 1.00 | 1 | 1 |
| ............................................................................ATTGCACTAGTCCCGGCCT.................................... | 19 | 1 | 1.00 | 1 | 1 |
| ..................................AGGCCGTGCCCAGTGCTTATT............................................................................ | 21 | 1 | 1.00 | 1 | 1 |
| ..............................................................................TGCACTAGTCCCGGCCTG................................... | 18 | 1 | 1.00 | 1 | 1 |
| .............................................................................TTGCACTAGTCCCGGCCTGC.................................. | 20 | 1 | 1.00 | 1 | 1 |