| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:21472213-21472337 + | TTTCAGCCGAA--------TATAAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATCTA-------------T----AAA |
| droSim1 | chr3R:21300457-21300581 + | TTTCAGCCGAA--------TAAAAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATCAA-------------T----AAA |
| droSec1 | super_13:258996-259120 + | TTTCAGCCGAA--------TAAAAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATCAA-------------T----CAA |
| droYak2 | chr3R:25001360-25001484 - | TTTCGTTCGAA--------TATGAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTTGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATGCA-------------T----TTT |
| droEre2 | scaffold_4820:6569995-6570123 - | TTTCGGCCGAA--------TATGAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTGAAATGAA-------------TTTTATAT |
| droAna3 | scaffold_13340:3727762-3727906 + | TTTCCATCGAA--------TAT-AAT----------------------------------------------ACAAATTTCCCGTAGGACGGGAAGGTGTCAAGGTTTTGAATT-AAATT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTCAA-----ACACATCGATACATATTTTTAG---AAATATTTAAAATTTGG----ATA |
| dp4 | chr2:23635253-23635419 - | CTTTA--------------------TATGCATATCCTGTAGGACAAGACTACAGTACTACTCGGAATACCATAGAGATTGCCCGTAGGACGGGAAGGTGTCAACGTTTTAAATTTTGAAC-AAACATTGCACTTGTCCCGGCCTATGGGCGGCTTGTCAT-----ACACATCGAT-CGTACAACTAG---CAA-------------C----AAA |
| droPer1 | super_3:6410293-6410459 - | CTTTA--------------------TATGCATATCCTGTAGGACAAGACTACAGTACTACTCGGAATACCATAGAGATTGCCCGTAGGACGGGAAGGTGTCAACGTTTTACATTTTGAAC-AAACATTGCACTTGTCCCGGCCTATGGGCGGCTTGTCAT-----ACACATCGAT-CGTACAACTAG---CAA-------------C----AAA |
| droWil1 | scaffold_181130:14578411-14578543 - | CTGTAGCCGGGTGAAACATT----ATCTGCAT----------------------------------------TCAAATTTCTCGTAGGACGGGAAGGTGTCAACGTTTTGCCTTATGAATCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTGTAAAAC----GACATC-----------CTAT---CA-------------------AAA |
| droVir3 | scaffold_12855:6880771-6880902 + | CGACAATATGG--------CTTTATC----------------------------------------------ACAAACTGCTTGCAGGACGGGAAGGTGTCAACGTTTTGCTTTATGAATCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTGTGCCACAACAAACG------------ATTGAAAATAT------------GA----AAA |
| droMoj3 | scaffold_6540:25734544-25734668 - | CTGCAAT--AC--------CCAAATC----------------------------------------------ATAAATTGCTTGCAGGACGGGAAGGTGTCAACGTTTTACCTTTTGATTCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTATCTTAC----AACA------------ATCAGGTCCAA-------------T----AGA |
| droGri2 | scaffold_14906:3023231-3023367 + | TTTTT---TAA--------TATATAT------ATTCT-TAA-AC-------------------GTGTACC--ATAAATTGCTTGCAGGACGGGAAGGTGTCAACGTTTTGCATTTTGAATCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTGTGCAT----CTACA------------ACTGGT----T-------------T----AAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:21472213-21472337 + | TTTCAGCCGAA--------TATAAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATCTA-------------T----AAA |
| droSim1 | chr3R:21300457-21300581 + | TTTCAGCCGAA--------TAAAAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATCAA-------------T----AAA |
| droSec1 | super_13:258996-259120 + | TTTCAGCCGAA--------TAAAAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATCAA-------------T----CAA |
| droYak2 | chr3R:25001360-25001484 - | TTTCGTTCGAA--------TATGAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTTGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTAAAATGCA-------------T----TTT |
| droEre2 | scaffold_4820:6569995-6570123 - | TTTCGGCCGAA--------TATGAAT----------------------------------------------ATGAATTTCCCGTAGGACGGGAAGGTGTCAACGTTTTGCATTTCGAAT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTAAT-----AAACA------------ACTGAAATGAA-------------TTTTATAT |
| droAna3 | scaffold_13340:3727762-3727906 + | TTTCCATCGAA--------TAT-AAT----------------------------------------------ACAAATTTCCCGTAGGACGGGAAGGTGTCAAGGTTTTGAATT-AAATT-AAACATTGCACTTGTCCCGGCCTATGGGCGGTTTGTCAA-----ACACATCGATACATATTTTTAG---AAATATTTAAAATTTGG----ATA |
| dp4 | chr2:23635253-23635419 - | CTTTA--------------------TATGCATATCCTGTAGGACAAGACTACAGTACTACTCGGAATACCATAGAGATTGCCCGTAGGACGGGAAGGTGTCAACGTTTTAAATTTTGAAC-AAACATTGCACTTGTCCCGGCCTATGGGCGGCTTGTCAT-----ACACATCGAT-CGTACAACTAG---CAA-------------C----AAA |
| droPer1 | super_3:6410293-6410459 - | CTTTA--------------------TATGCATATCCTGTAGGACAAGACTACAGTACTACTCGGAATACCATAGAGATTGCCCGTAGGACGGGAAGGTGTCAACGTTTTACATTTTGAAC-AAACATTGCACTTGTCCCGGCCTATGGGCGGCTTGTCAT-----ACACATCGAT-CGTACAACTAG---CAA-------------C----AAA |
| droWil1 | scaffold_181130:14578411-14578543 - | CTGTAGCCGGGTGAAACATT----ATCTGCAT----------------------------------------TCAAATTTCTCGTAGGACGGGAAGGTGTCAACGTTTTGCCTTATGAATCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTGTAAAAC----GACATC-----------CTAT---CA-------------------AAA |
| droVir3 | scaffold_12855:6880771-6880902 + | CGACAATATGG--------CTTTATC----------------------------------------------ACAAACTGCTTGCAGGACGGGAAGGTGTCAACGTTTTGCTTTATGAATCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTGTGCCACAACAAACG------------ATTGAAAATAT------------GA----AAA |
| droMoj3 | scaffold_6540:25734544-25734668 - | CTGCAAT--AC--------CCAAATC----------------------------------------------ATAAATTGCTTGCAGGACGGGAAGGTGTCAACGTTTTACCTTTTGATTCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTATCTTAC----AACA------------ATCAGGTCCAA-------------T----AGA |
| droGri2 | scaffold_14906:3023231-3023367 + | TTTTT---TAA--------TATATAT------ATTCT-TAA-AC-------------------GTGTACC--ATAAATTGCTTGCAGGACGGGAAGGTGTCAACGTTTTGCATTTTGAATCAAACATTGCACTTGTCCCGGCCTATGAGCGGTTTGTGCAT----CTACA------------ACTGGT----T-------------T----AAA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-31.5, p-value=0.009901 |
|---|
 |
| dG=-31.5, p-value=0.009901 |
|---|
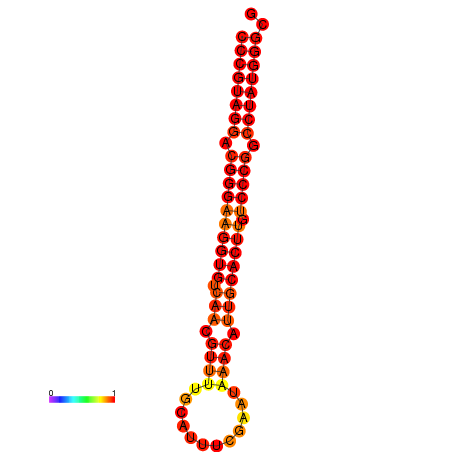
 |
| dG=-31.5, p-value=0.009901 |
|---|
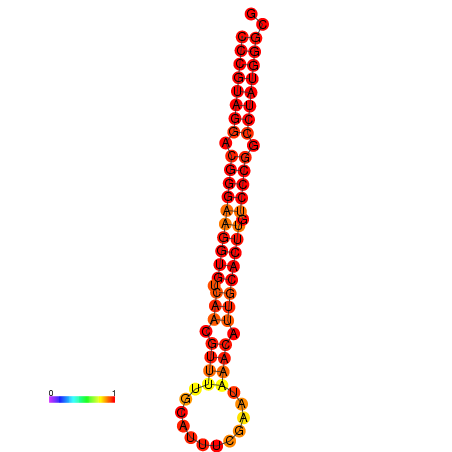
 |
| dG=-31.5, p-value=0.009901 | dG=-30.5, p-value=0.009901 |
|---|---|
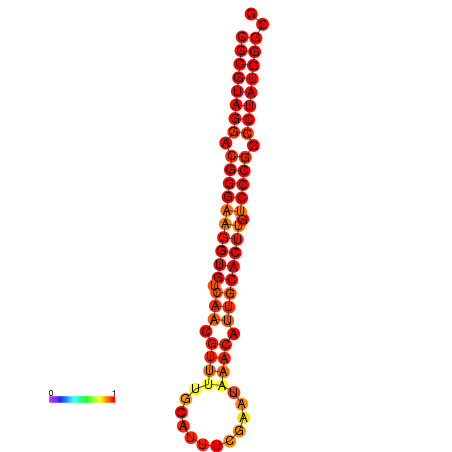
 |
 |
| dG=-31.5, p-value=0.009901 |
|---|
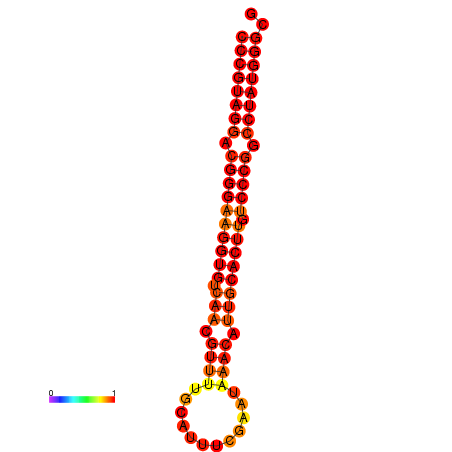 |
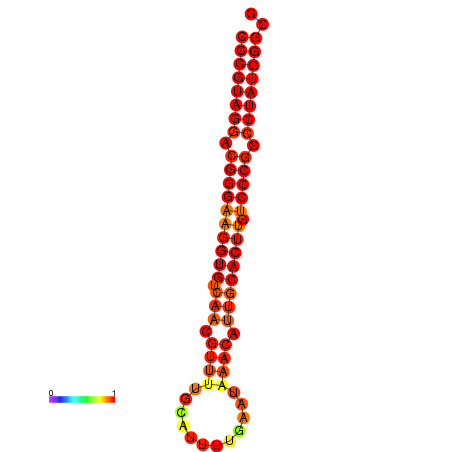
| dG=-31.6, p-value=0.009901 | dG=-31.0, p-value=0.009901 | dG=-30.7, p-value=0.009901 | dG=-30.7, p-value=0.009901 | dG=-30.7, p-value=0.009901 |
|---|---|---|---|---|
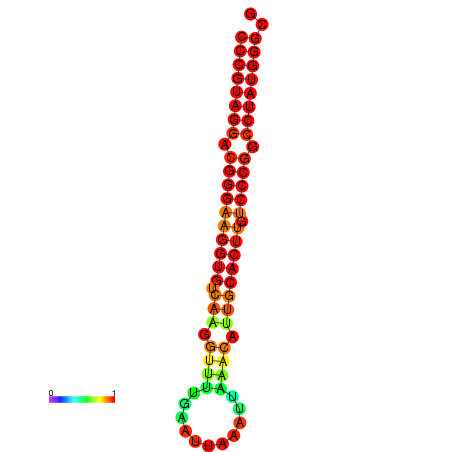 |
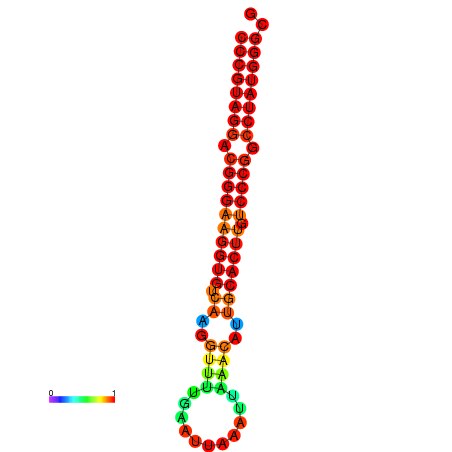 |
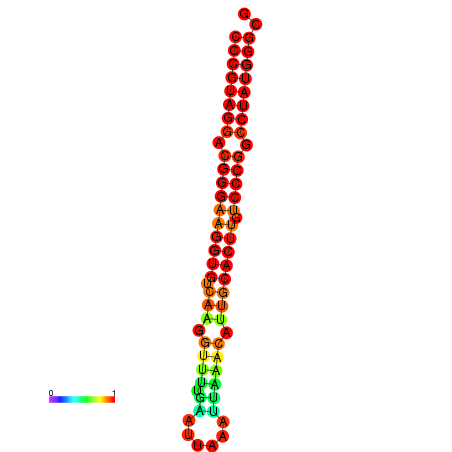 |
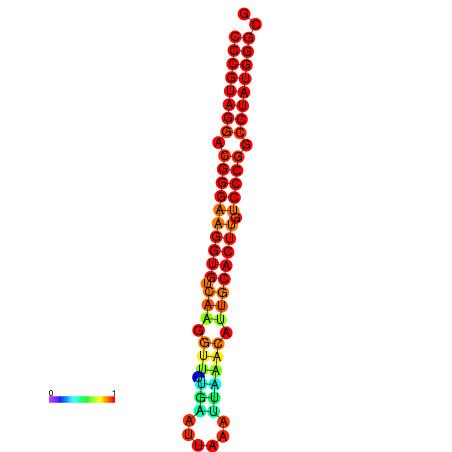 |
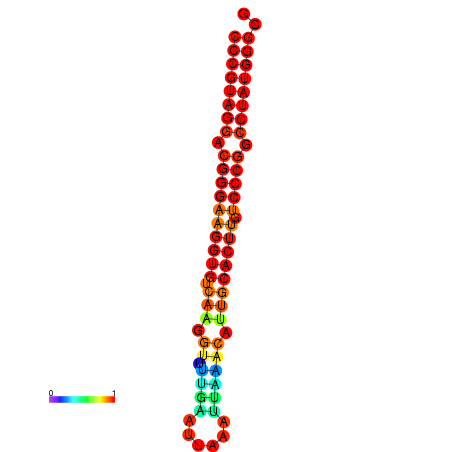 |
| dG=-30.7, p-value=0.009901 | dG=-29.7, p-value=0.009901 |
|---|---|
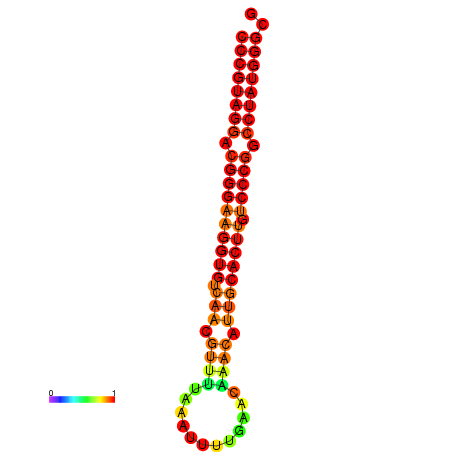 |
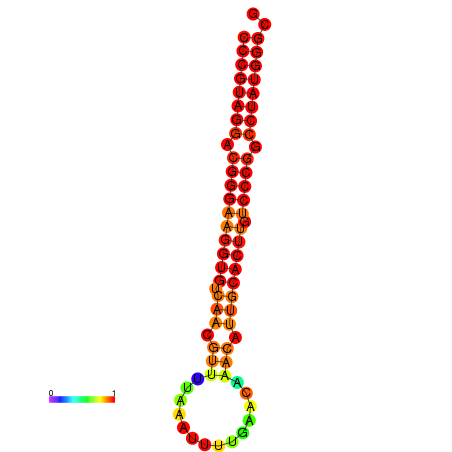 |
| dG=-30.7, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-29.7, p-value=0.009901 |
|---|---|---|
 |
 |
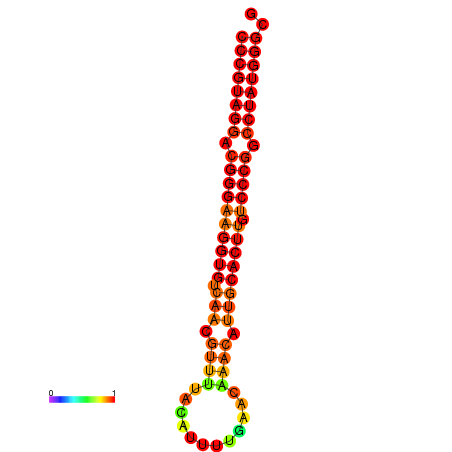
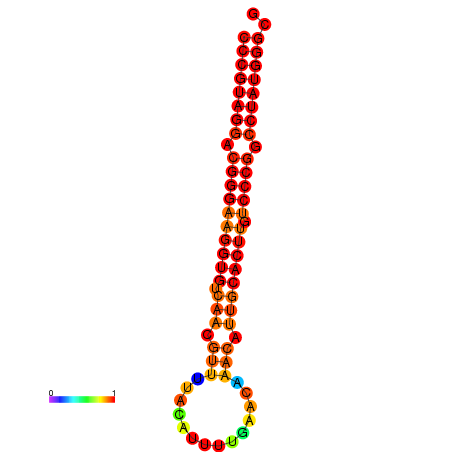 |
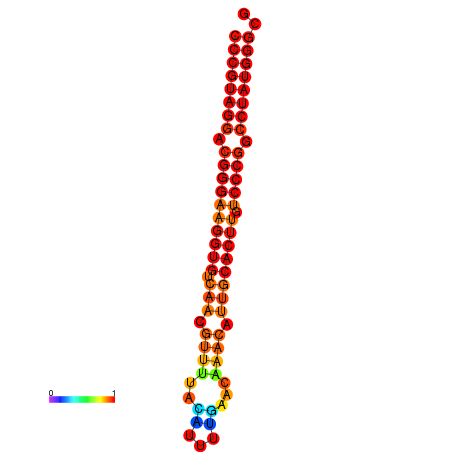
| dG=-28.5, p-value=0.009901 | dG=-27.6, p-value=0.009901 | dG=-27.5, p-value=0.009901 | dG=-27.5, p-value=0.009901 |
|---|---|---|---|
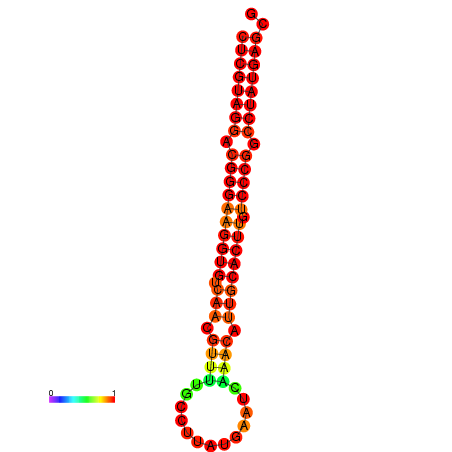 |
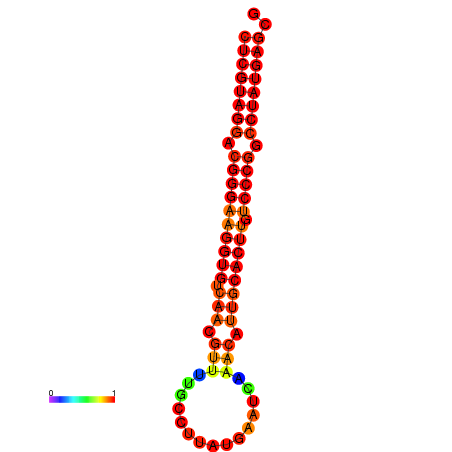 |
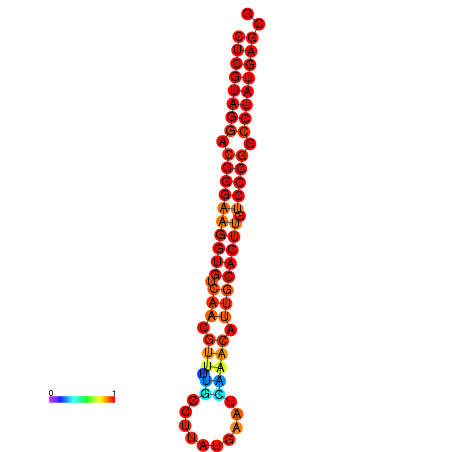 |
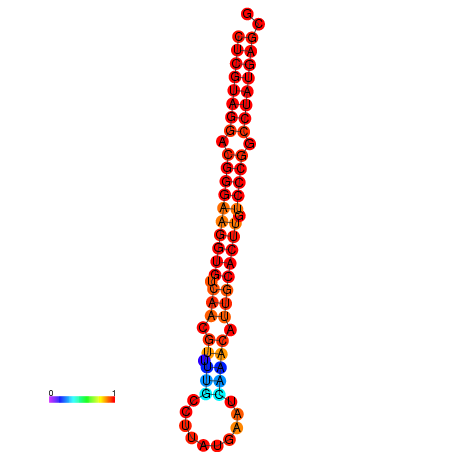 |
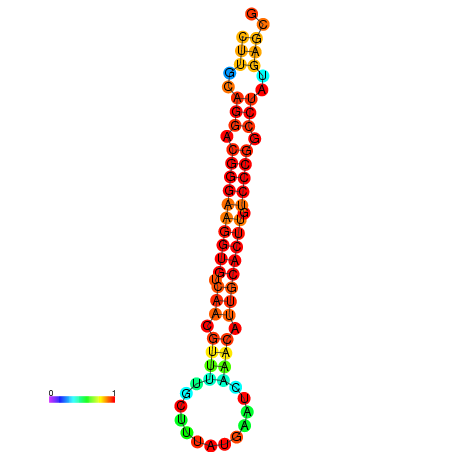
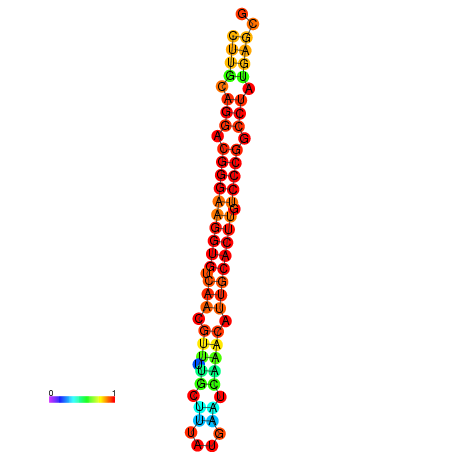
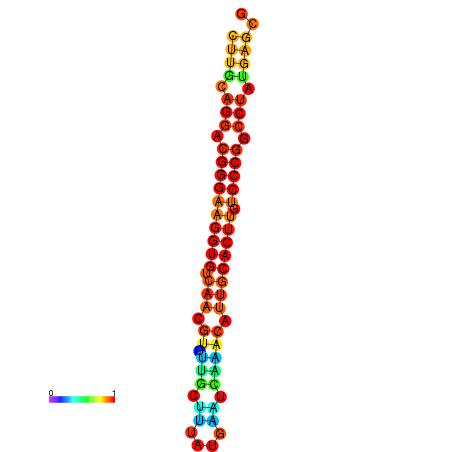
| dG=-21.1, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|---|
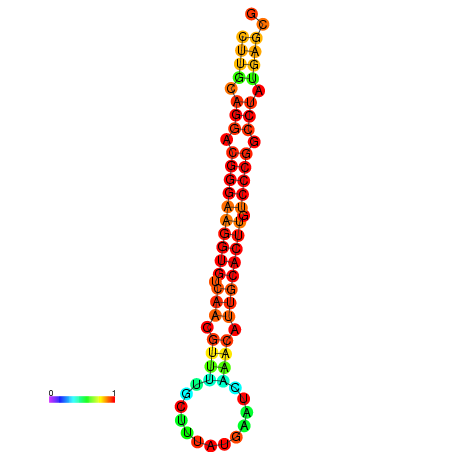 |
 |
 |
 |
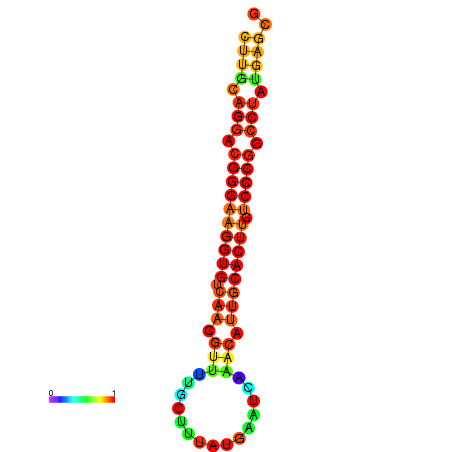 |
| dG=-21.1, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|
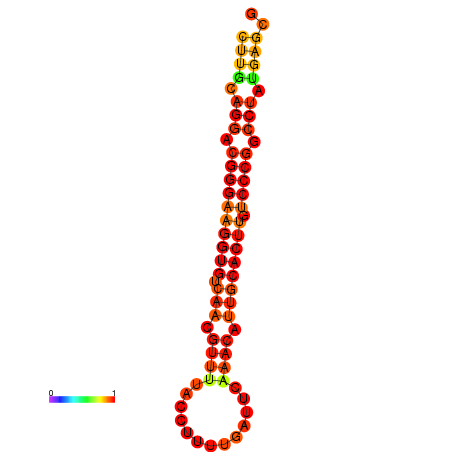 |
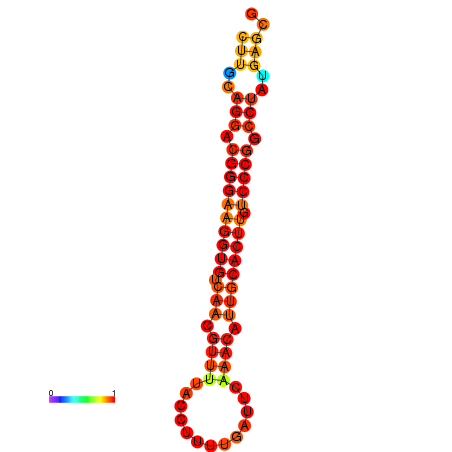 |
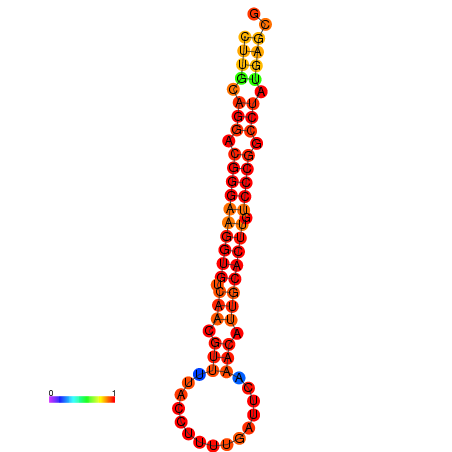 |
| dG=-21.1, p-value=0.009901 | dG=-20.9, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.5, p-value=0.009901 |
|---|---|---|---|---|
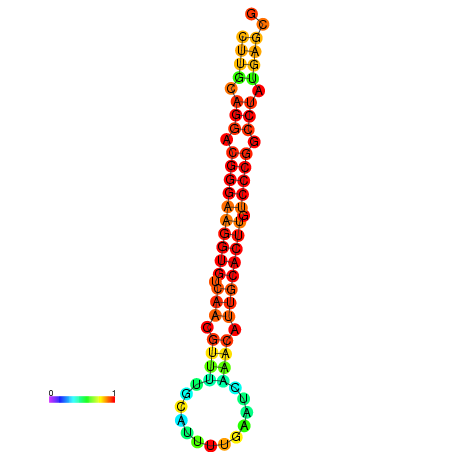 |
 |
 |
 |
 |
Generated: 03/07/2013 at 07:03 PM