| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:121076-121193 + | TGA--C-ATGGCG---GAACCAG-------------TCCTGGTGGAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAGCGACTCCCTAACGGAGTCAGATTGAGCTGCAAAGGAGCGA-CACAAC-----AGAGGAGC----A |
| droSim1 | chr3R:194547-194664 + | TGG--C-ATGGCG---CAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAGCGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----AGAGGAGC----A |
| droSec1 | super_6:240419-240536 + | TGA--C-ATGGCG---CAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAGCGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----AGAGGAGC----A |
| droYak2 | chr3R:481089-481206 + | TGA--A-ATGGCA---AAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAACGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----ACAGGACC----A |
| droEre2 | scaffold_4770:471031-471148 + | TGA--A-GTGGCG---GAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTC-CATAAGCGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----ACAGGTGC----A |
| droAna3 | scaffold_13340:11127706-11127842 + | AGC--C-ACCAAC---CAACCAACCAACCAACCACTTCCTGGCGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTAACGAGAGACTCCCTAACGGAGTCAGATTGAATTACAAAGGAGCGA-GACAACGACTACGTGGTGG----C |
| dp4 | chr2:24810831-24810947 - | G-A--A-GT-GCA---CGGCAA-------------TTCCTGCAGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTATTGAGCGACTCCCTAACGGAGTCAGATTGAATTACCAAGGAAC-A-GACAAC-----AGTGGAGC----G |
| droPer1 | super_6:111437-111553 - | G-A--A-GT-GCA---CGG--G-----------ATTTCCTGCAGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTATTGAGCGACTCCCTAACGGAGTCAGATTGAATTACCAAGGAAC-A-GACAAC-----AGTGGAGC----A |
| droWil1 | scaffold_181089:9825565-9825678 - | TGA-------------GAA---------------CTTCCTGCGGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTATATAAGCGACTCCCTAACGGAGTCAGATTGAATTACAAAGGAACAACAACAAC-----AAAAA-GCCACGA |
| droVir3 | scaffold_12822:1418795-1418911 + | AGGCCAAGT-GCGACGCGT--G-----------AATTCCTGCCGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTT-CACGAGCGACTCCCTAACGGAGTCAGATTGAATTACAAAGGACCCA-GACAAC-----GAG---------- |
| droMoj3 | scaffold_6540:34020273-34020392 + | ACA--T-AT-ACG---CTACGAG-----------TTTCCTCATGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTTTTTAACGACTCCCTAACGGAGTCAGATTGAATTTCAGAGGACCCA-GACAAC-----GAAGAAGC----G |
| droGri2 | scaffold_15074:2468173-2468294 - | TGA--AAGT-GTGCCCCCG--A-----------ACTTCCTGCTGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTAACGAGCGACTCCCTAACGGAGTCAGATTGAGTTACAAAGGACCAA-GTCAAC-----AATGCCCC----A |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:121076-121193 + | TGA--C-ATGGCG---GAACCAG-------------TCCTGGTGGAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAGCGACTCCCTAACGGAGTCAGATTGAGCTGCAAAGGAGCGA-CACAAC-----AGAGGAGC----A |
| droSim1 | chr3R:194547-194664 + | TGG--C-ATGGCG---CAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAGCGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----AGAGGAGC----A |
| droSec1 | super_6:240419-240536 + | TGA--C-ATGGCG---CAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAGCGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----AGAGGAGC----A |
| droYak2 | chr3R:481089-481206 + | TGA--A-ATGGCA---AAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTT-AATGAACGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----ACAGGACC----A |
| droEre2 | scaffold_4770:471031-471148 + | TGA--A-GTGGCG---GAACCAG-------------TCCTGGTGAAGCTCAAATTGACTCTAGTAGGGAGTCCTTC-CATAAGCGACTCCCTAACGGAGTCAGATTGAGCTACAAAGGAGCGA-GACAAC-----ACAGGTGC----A |
| droAna3 | scaffold_13340:11127706-11127842 + | AGC--C-ACCAAC---CAACCAACCAACCAACCACTTCCTGGCGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTAACGAGAGACTCCCTAACGGAGTCAGATTGAATTACAAAGGAGCGA-GACAACGACTACGTGGTGG----C |
| dp4 | chr2:24810831-24810947 - | G-A--A-GT-GCA---CGGCAA-------------TTCCTGCAGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTATTGAGCGACTCCCTAACGGAGTCAGATTGAATTACCAAGGAAC-A-GACAAC-----AGTGGAGC----G |
| droPer1 | super_6:111437-111553 - | G-A--A-GT-GCA---CGG--G-----------ATTTCCTGCAGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTATTGAGCGACTCCCTAACGGAGTCAGATTGAATTACCAAGGAAC-A-GACAAC-----AGTGGAGC----A |
| droWil1 | scaffold_181089:9825565-9825678 - | TGA-------------GAA---------------CTTCCTGCGGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTATATAAGCGACTCCCTAACGGAGTCAGATTGAATTACAAAGGAACAACAACAAC-----AAAAA-GCCACGA |
| droVir3 | scaffold_12822:1418795-1418911 + | AGGCCAAGT-GCGACGCGT--G-----------AATTCCTGCCGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTT-CACGAGCGACTCCCTAACGGAGTCAGATTGAATTACAAAGGACCCA-GACAAC-----GAG---------- |
| droMoj3 | scaffold_6540:34020273-34020392 + | ACA--T-AT-ACG---CTACGAG-----------TTTCCTCATGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTTTTTAACGACTCCCTAACGGAGTCAGATTGAATTTCAGAGGACCCA-GACAAC-----GAAGAAGC----G |
| droGri2 | scaffold_15074:2468173-2468294 - | TGA--AAGT-GTGCCCCCG--A-----------ACTTCCTGCTGCAGTTCAAATTGACTCTAGTAGGGAGTCCTTTTAACGAGCGACTCCCTAACGGAGTCAGATTGAGTTACAAAGGACCAA-GTCAAC-----AATGCCCC----A |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-40.4, p-value=0.009901 | dG=-39.5, p-value=0.009901 |
|---|---|
 |
 |
| dG=-40.4, p-value=0.009901 | dG=-39.5, p-value=0.009901 |
|---|---|
 |
 |
| dG=-40.4, p-value=0.009901 | dG=-39.5, p-value=0.009901 |
|---|---|
 |
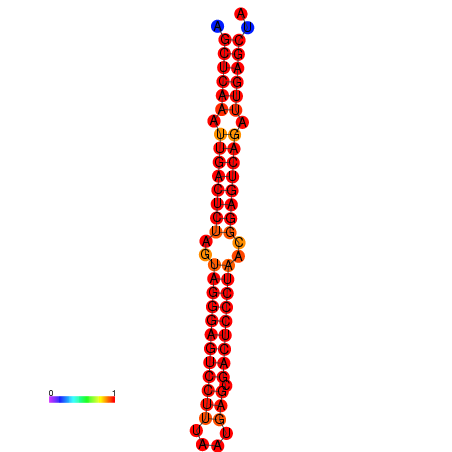 |
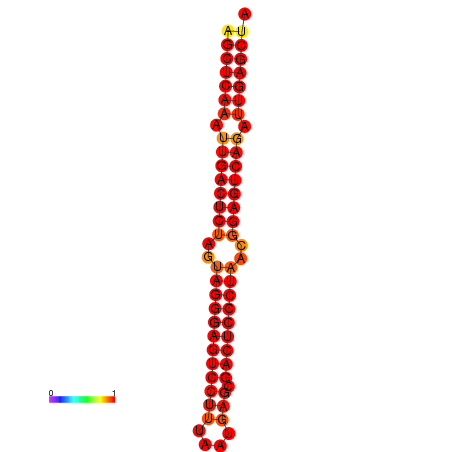
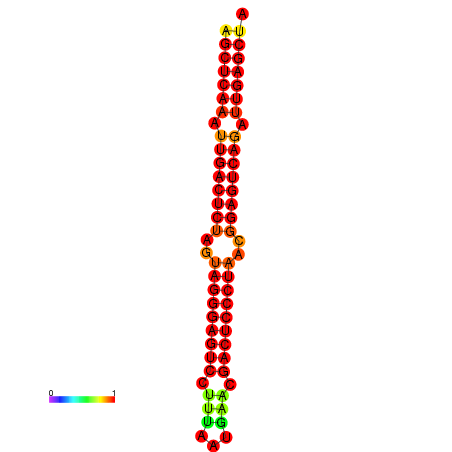
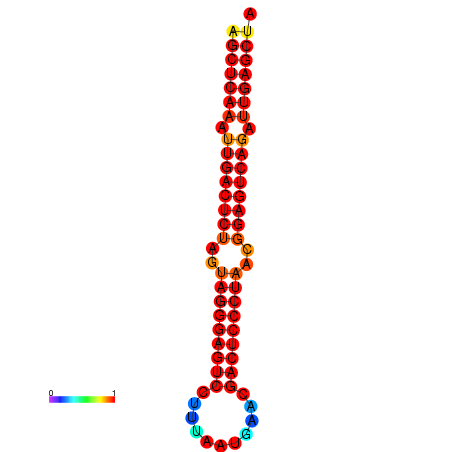
| dG=-37.3, p-value=0.009901 | dG=-36.7, p-value=0.009901 | dG=-36.4, p-value=0.009901 | dG=-36.4, p-value=0.009901 |
|---|---|---|---|
 |
 |
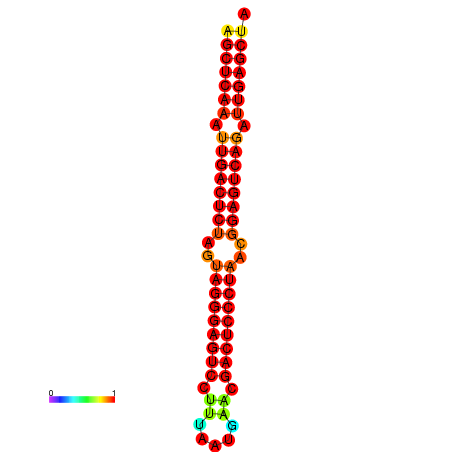 |
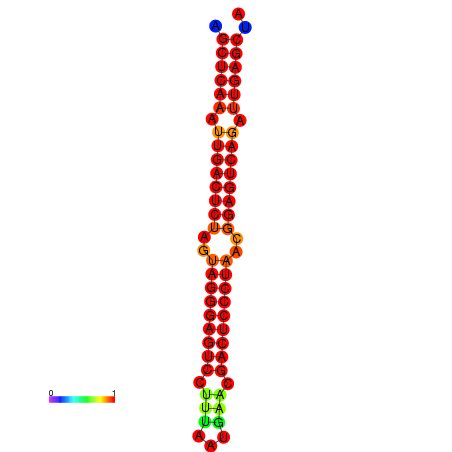 |
| dG=-39.1, p-value=0.009901 | dG=-38.6, p-value=0.009901 | dG=-38.2, p-value=0.009901 |
|---|---|---|
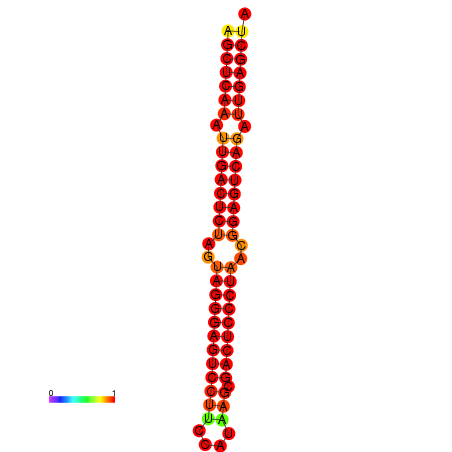 |
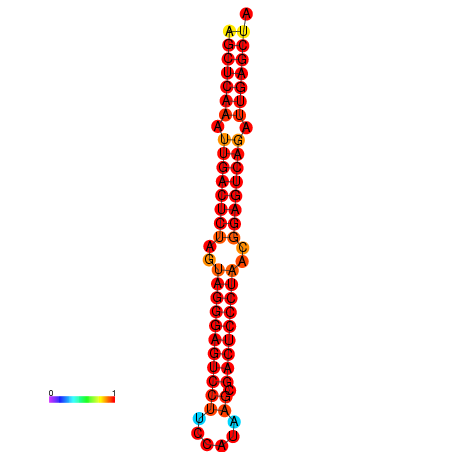 |
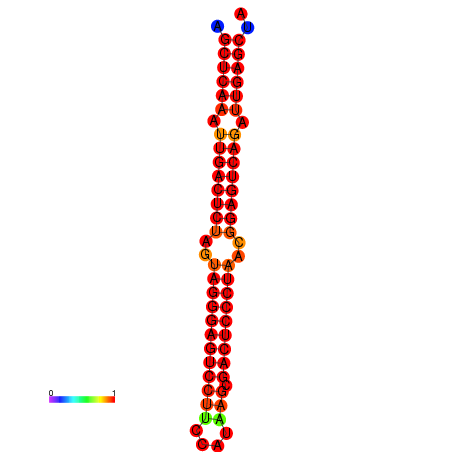 |
| dG=-34.8, p-value=0.009901 | dG=-34.4, p-value=0.009901 |
|---|---|
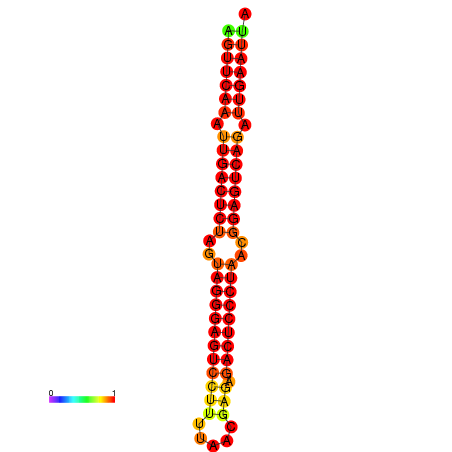 |
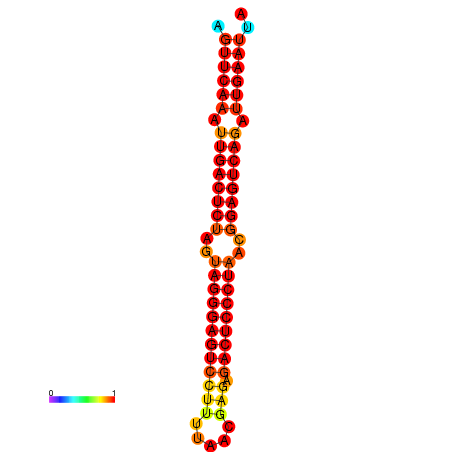 |
| dG=-35.6, p-value=0.009901 | dG=-35.2, p-value=0.009901 |
|---|---|
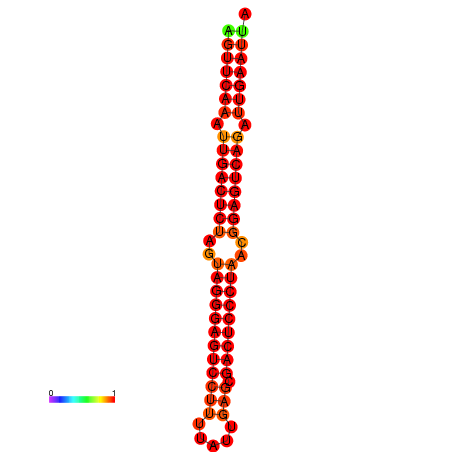 |
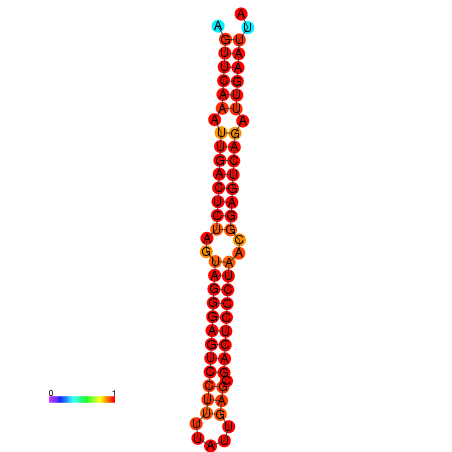 |
| dG=-35.6, p-value=0.009901 | dG=-35.2, p-value=0.009901 |
|---|---|
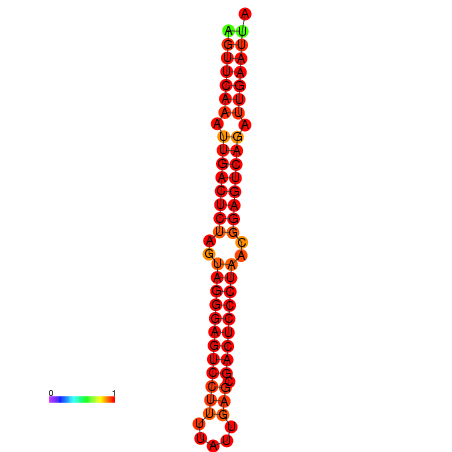 |
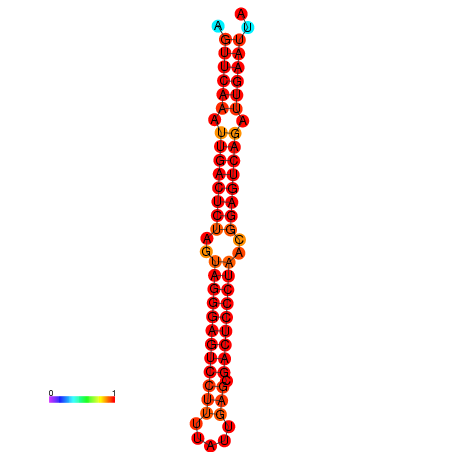 |
| dG=-35.2, p-value=0.009901 | dG=-34.8, p-value=0.009901 |
|---|---|
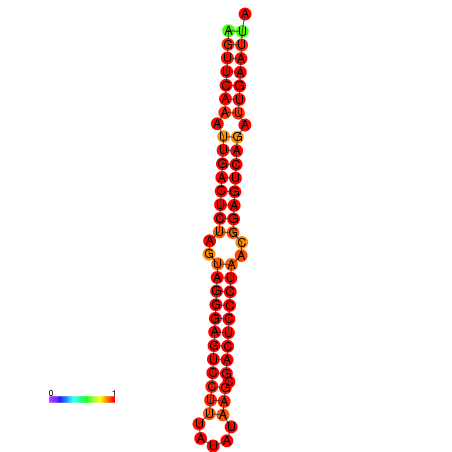 |
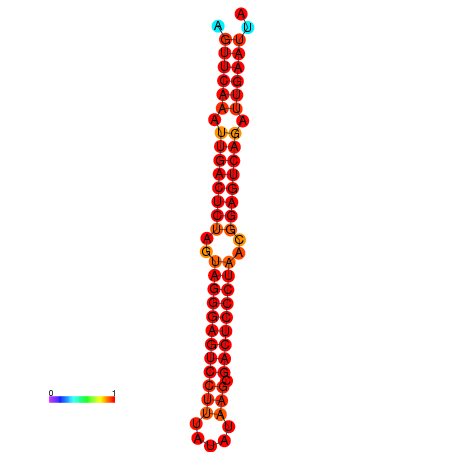 |
| dG=-34.9, p-value=0.009901 | dG=-34.5, p-value=0.009901 | dG=-33.9, p-value=0.009901 |
|---|---|---|
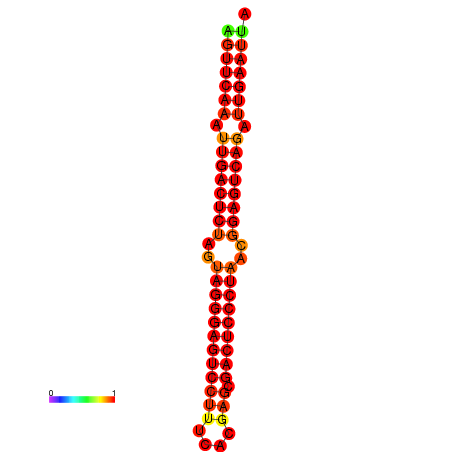 |
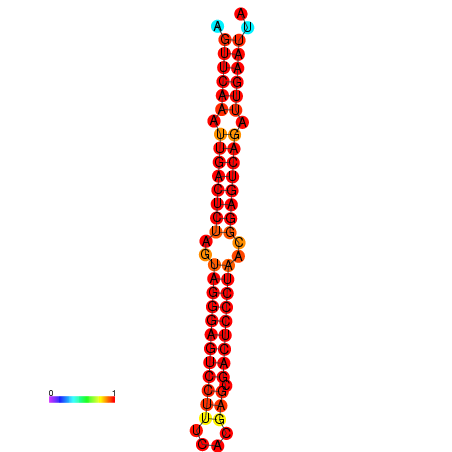 |
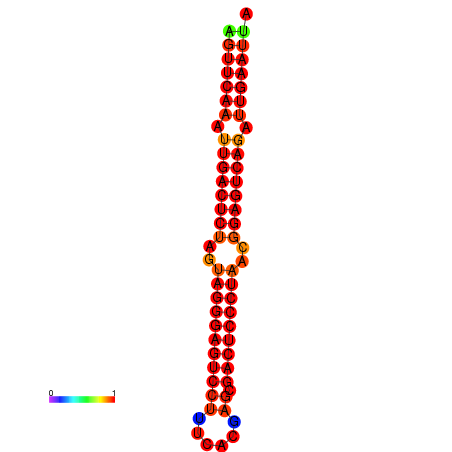 |
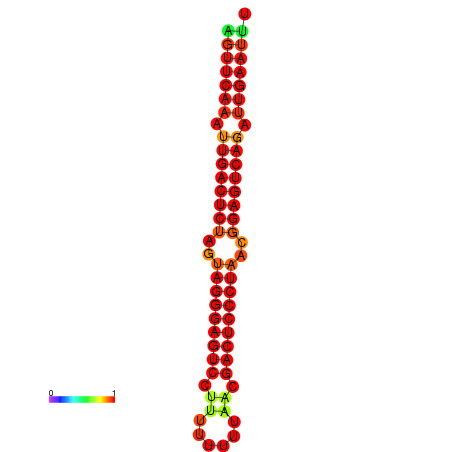
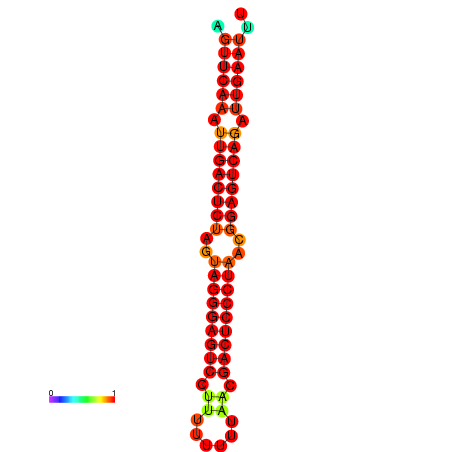
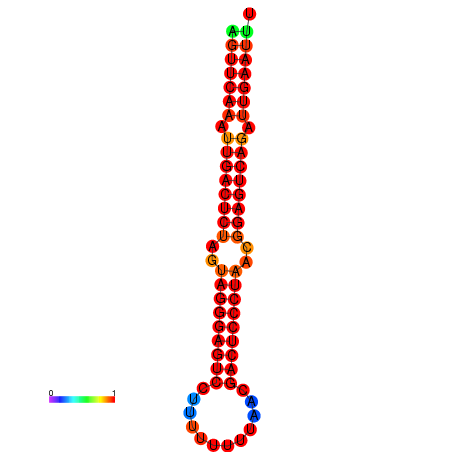
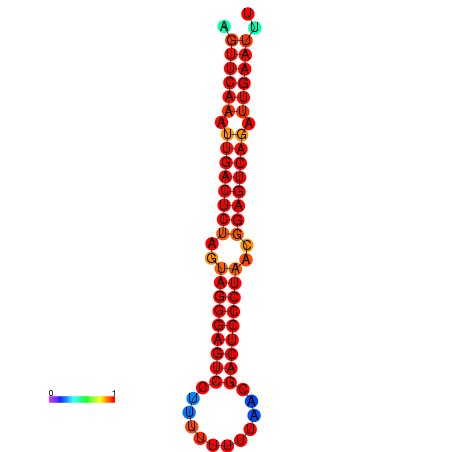
| dG=-32.5, p-value=0.009901 | dG=-32.3, p-value=0.009901 | dG=-31.7, p-value=0.009901 | dG=-31.5, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
 |
| dG=-32.5, p-value=0.009901 | dG=-31.8, p-value=0.009901 | dG=-31.8, p-value=0.009901 |
|---|---|---|
 |
 |
 |
Generated: 03/07/2013 at 07:03 PM