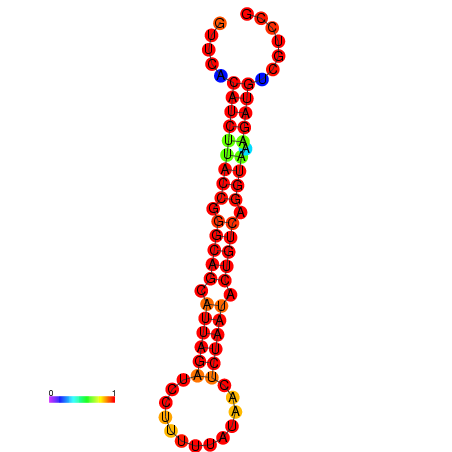
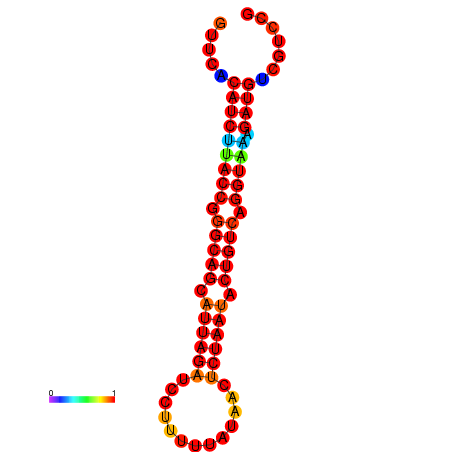
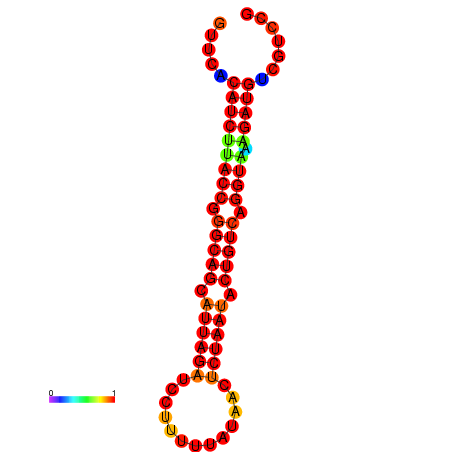
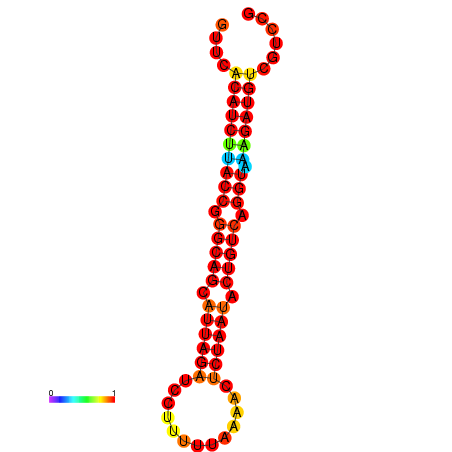
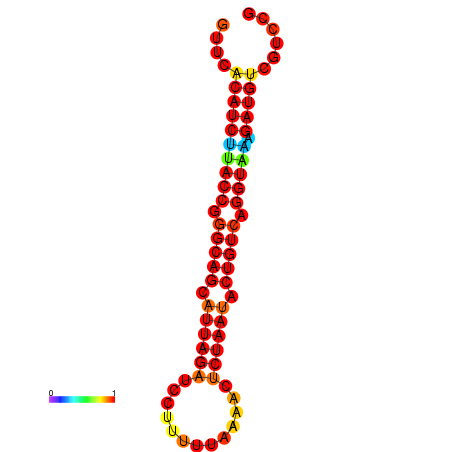
| AAATAACAAAAGAGGGAAACACAGTGCGGTGACGAAAACACGTTTTGCTA-TTCGCAAGGACATCTGTTCACATCTTACCGGGCAGCATTAGATCCTTTAGATACCTCTAATACTGTCAGGTAAAGATGTCGTCCGTGTCCTTATCCATCATAAATCC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ............................................................................................................TAATACTGTCAGGTAAAGATGTC........................... | 23 | 1 | 447032.00 | 447032 | 198408 | 248624 |
| ............................................................................................................TAATACTGTCAGGTAAAGATGT............................ | 22 | 1 | 69948.00 | 69948 | 8448 | 61500 |
| ............................................................................................................TAATACTGTCAGGTAAAGATG............................. | 21 | 1 | 47189.00 | 47189 | 5844 | 41345 |
| .......................................................................CATCTTACCGGGCAGCATTAGA................................................................. | 22 | 1 | 11663.00 | 11663 | 5614 | 6049 |
| .............................................................................................................AATACTGTCAGGTAAAGATGTC........................... | 22 | 1 | 1371.00 | 1371 | 245 | 1126 |
| .......................................................................CATCTTACCGGGCAGCATTAG.................................................................. | 21 | 1 | 1277.00 | 1277 | 625 | 652 |
| ............................................................................................................TAATACTGTCAGGTAAAGAT.............................. | 20 | 1 | 1100.00 | 1100 | 63 | 1037 |
| ............................................................................................................TAATACTGTCAGGTAAAGA............................... | 19 | 1 | 432.00 | 432 | 51 | 381 |
| ...........................................................................................................CTAATACTGTCAGGTAAAGATGTC........................... | 24 | 1 | 274.00 | 274 | 136 | 138 |
| ..............................................................................................................ATACTGTCAGGTAAAGATGTC........................... | 21 | 1 | 247.00 | 247 | 79 | 168 |
| .............................................................................................................AATACTGTCAGGTAAAGATGT............................ | 21 | 1 | 166.00 | 166 | 16 | 150 |
| ...............................................................................................................TACTGTCAGGTAAAGATGTC........................... | 20 | 1 | 143.00 | 143 | 22 | 121 |
| ........................................................................ATCTTACCGGGCAGCATTAGA................................................................. | 21 | 1 | 118.00 | 118 | 45 | 73 |
| .......................................................................CATCTTACCGGGCAGCATTA................................................................... | 20 | 1 | 117.00 | 117 | 15 | 102 |
| ............................................................................................................TAATACTGTCAGGTAAAGATGTCG.......................... | 24 | 1 | 116.00 | 116 | 46 | 70 |
| ................................................................................................................ACTGTCAGGTAAAGATGTC........................... | 19 | 1 | 96.00 | 96 | 34 | 62 |
| .......................................................................CATCTTACCGGGCAGCATT.................................................................... | 19 | 1 | 74.00 | 74 | 11 | 63 |
| .................................................................................................................CTGTCAGGTAAAGATGTC........................... | 18 | 1 | 68.00 | 68 | 32 | 36 |
| ...........................................................................................................CTAATACTGTCAGGTAAAGATGT............................ | 23 | 1 | 52.00 | 52 | 5 | 47 |
| .............................................................................................................AATACTGTCAGGTAAAGATG............................. | 20 | 1 | 46.00 | 46 | 4 | 42 |
| ..............................................................................................................ATACTGTCAGGTAAAGATGT............................ | 20 | 1 | 41.00 | 41 | 2 | 39 |