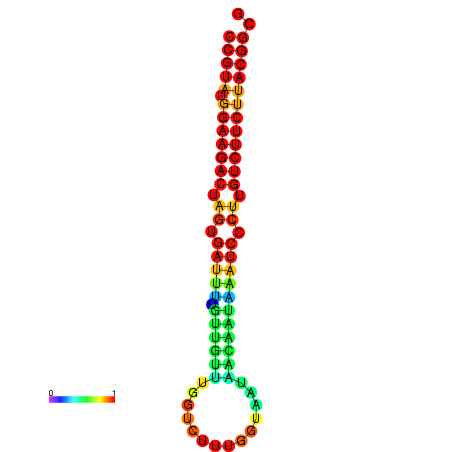
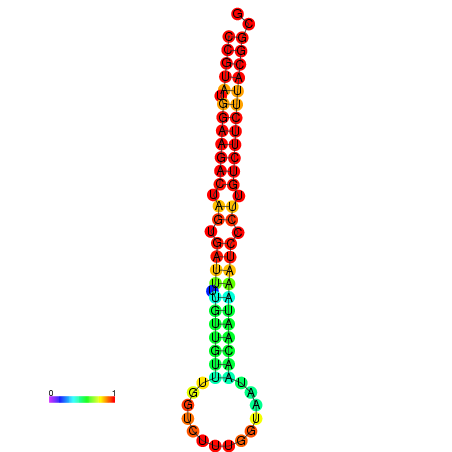
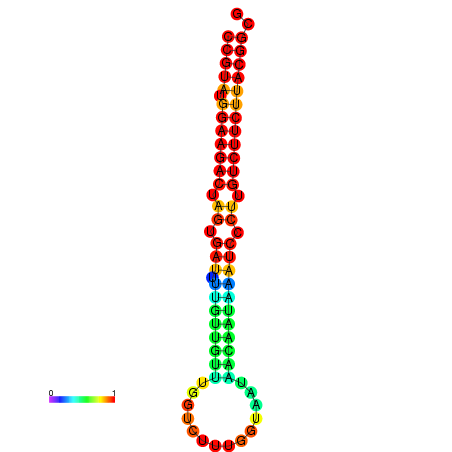
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16493557-16493674 + | GATTGGTC-CTCC-TGGGAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droSim1 | chr2R:15154934-15155051 + | GATTGGTC-CTCC-TGGGAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droSec1 | super_1:14047859-14047976 + | GATTGGTC-CTCC-TGGGAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droYak2 | chr2R:12702727-12702844 - | GACTGGTC-CTCC-TGGGAGTGCATTTCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droEre2 | scaffold_4845:10640174-10640291 + | GATTGGTC-CTCC-TGGGAGTGTATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droAna3 | scaffold_13266:4109353-4109468 - | TGGGAGTC-CT-G-TGGAAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTTTATGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTCG--GCTCATCACTCT----------A |
| dp4 | chr3:8655644-8655762 - | GATTGGCCTCTCC-TGGGAGTGTATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATCTG-TGCTCTTCATTCT----------A |
| droPer1 | super_4:2540080-2540198 - | GATTGGCCTCTCC-TGGGAGTGTATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATCTG-TGCTCTTCATTCT----------A |
| droWil1 | scaffold_180700:3423572-3423692 - | AAATTGTGTGTGG-T-GGAATGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTAATAAAATTAACAATAAATCCCTTGTCTTCTTATGGCGTGTATTTAAAGCTCTTCATT-T----------G |
| droVir3 | scaffold_12823:2295157-2295274 - | G--TCGT-----C-TCCAAATGCTTTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTATGGCGCGCAT-TC-TGCTCTTCATTTATAATA--A-TT |
| droMoj3 | scaffold_6496:1650020-1650148 - | TAACCGTTCGTCGTATCGAGTGCTTGCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTATGGCTCGCAT-TG-AGCTCTTCATTTCAAATCTCGTTA |
| droGri2 | scaffold_15245:15782213-15782323 + | GTTT-------------AAATGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGTTAA---TAACAATAAATCCCTTGTCTCCTAACGGCGCGCAT-TG-TGCTCTTCATTAT----TTC-TTA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:16493557-16493674 + | GATTGGTC-CTCC-TGGGAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droSim1 | chr2R:15154934-15155051 + | GATTGGTC-CTCC-TGGGAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droSec1 | super_1:14047859-14047976 + | GATTGGTC-CTCC-TGGGAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droYak2 | chr2R:12702727-12702844 - | GACTGGTC-CTCC-TGGGAGTGCATTTCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droEre2 | scaffold_4845:10640174-10640291 + | GATTGGTC-CTCC-TGGGAGTGTATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTTG-TGCTCTTCATTCT----------A |
| droAna3 | scaffold_13266:4109353-4109468 - | TGGGAGTC-CT-G-TGGAAGTGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTTTATGGTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATTCG--GCTCATCACTCT----------A |
| dp4 | chr3:8655644-8655762 - | GATTGGCCTCTCC-TGGGAGTGTATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATCTG-TGCTCTTCATTCT----------A |
| droPer1 | super_4:2540080-2540198 - | GATTGGCCTCTCC-TGGGAGTGTATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTACGGCGTGCATCTG-TGCTCTTCATTCT----------A |
| droWil1 | scaffold_180700:3423572-3423692 - | AAATTGTGTGTGG-T-GGAATGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTAATAAAATTAACAATAAATCCCTTGTCTTCTTATGGCGTGTATTTAAAGCTCTTCATT-T----------G |
| droVir3 | scaffold_12823:2295157-2295274 - | G--TCGT-----C-TCCAAATGCTTTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTATGGCGCGCAT-TC-TGCTCTTCATTTATAATA--A-TT |
| droMoj3 | scaffold_6496:1650020-1650148 - | TAACCGTTCGTCGTATCGAGTGCTTGCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTCTGCTAA---TAACAATAAATCCCTTGTCTTCTTATGGCTCGCAT-TG-AGCTCTTCATTTCAAATCTCGTTA |
| droGri2 | scaffold_15245:15782213-15782323 + | GTTT-------------AAATGCATTCCGTATGGAAGACTAGTGATTTTGTTGTTTGGTCTTTGTTAA---TAACAATAAATCCCTTGTCTCCTAACGGCGCGCAT-TG-TGCTCTTCATTAT----TTC-TTA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
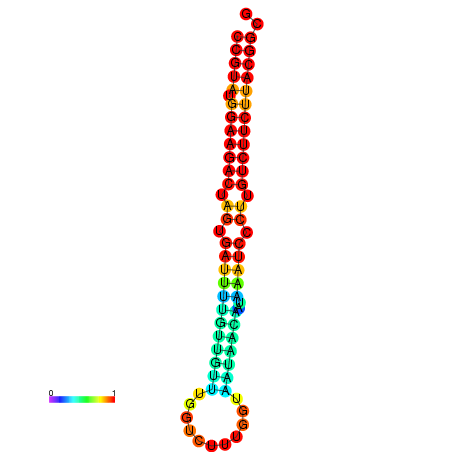
| dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
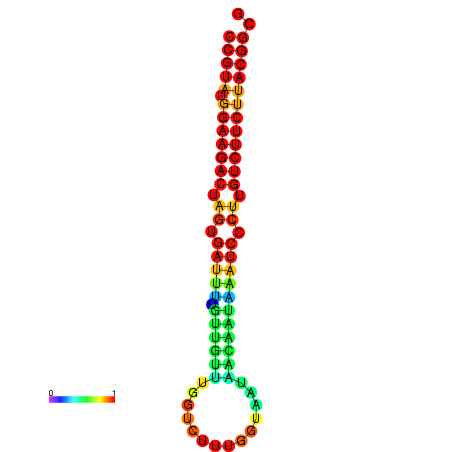 |
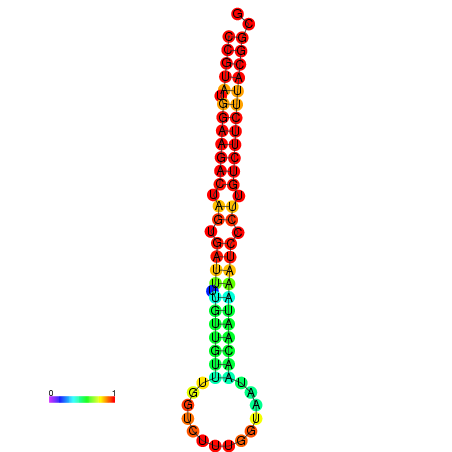 |
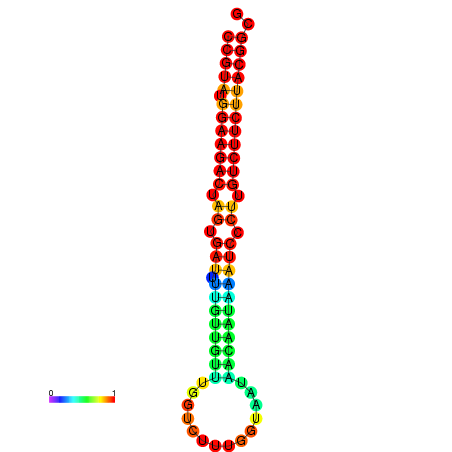 |
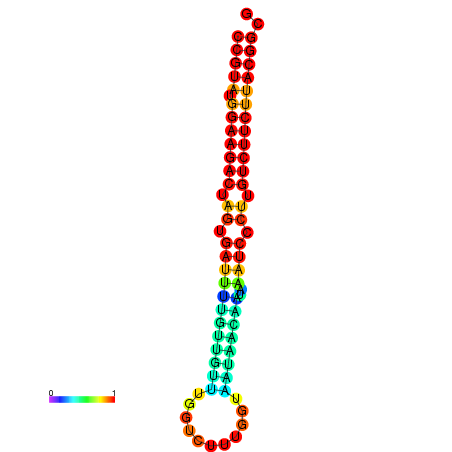
| dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|---|
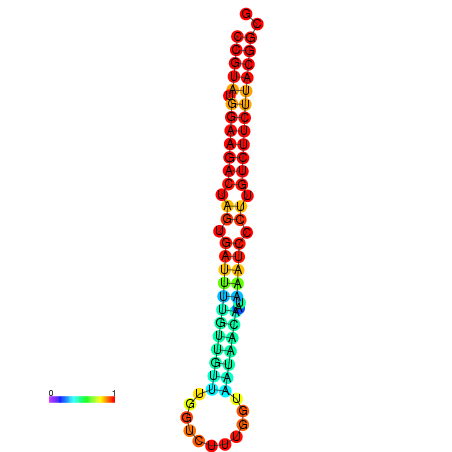 |
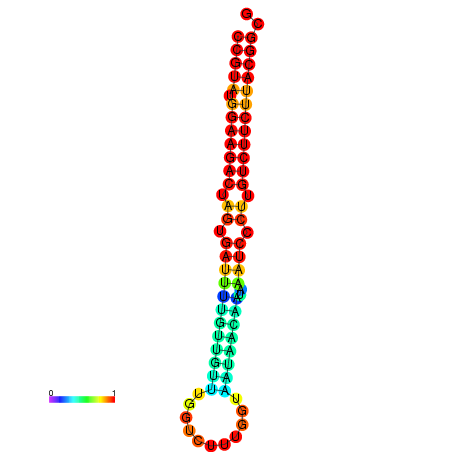 |
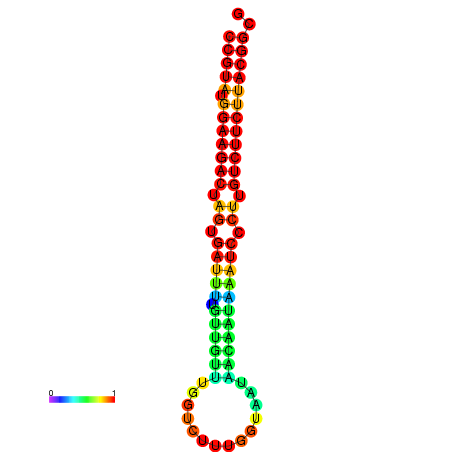 |
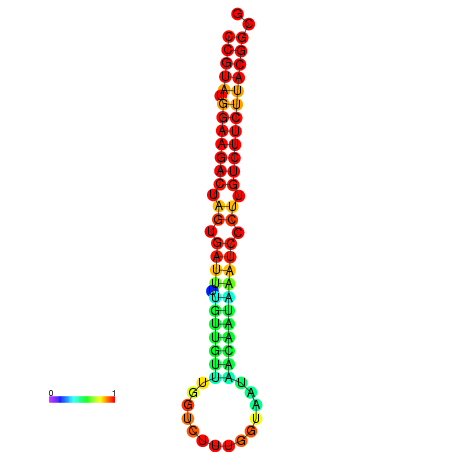 |
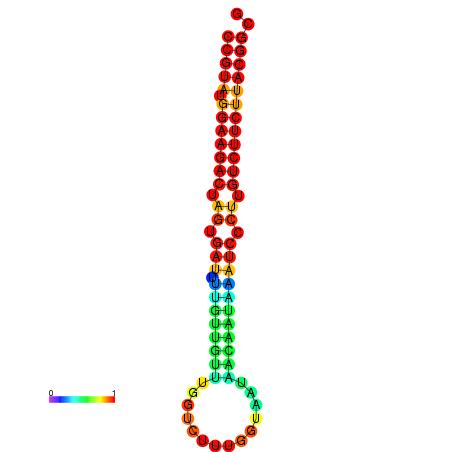 |
| dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|---|
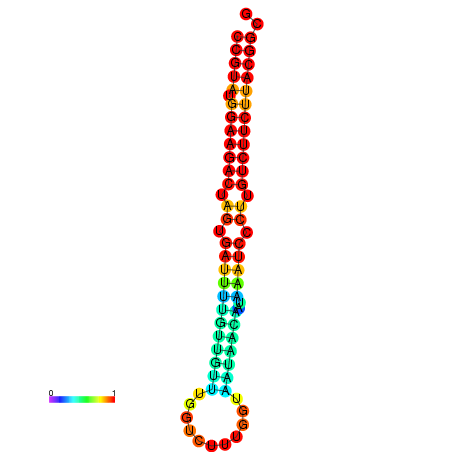 |
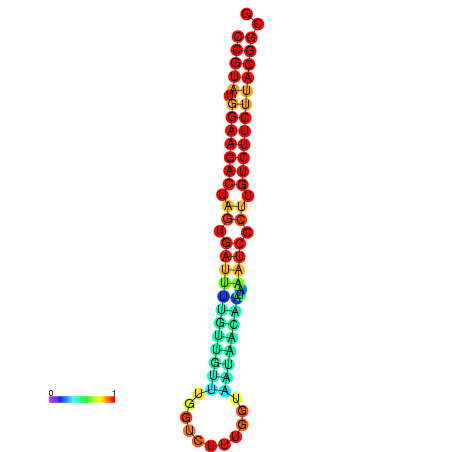 |
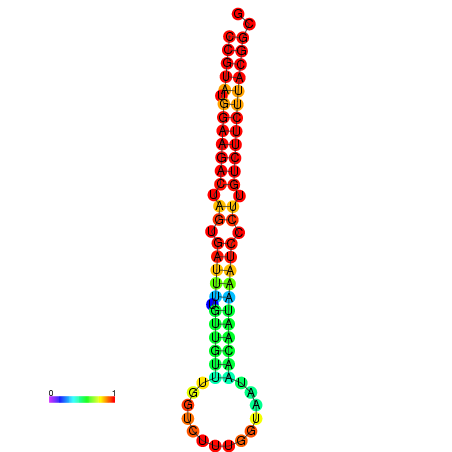 |
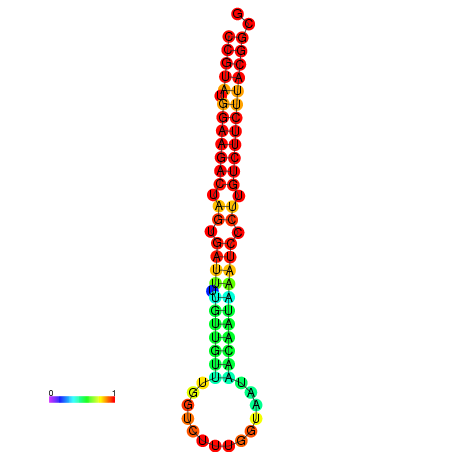 |
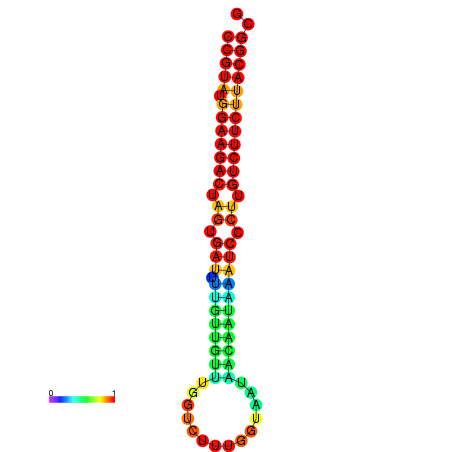 |
| dG=-20.5, p-value=0.009901 | dG=-20.5, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.2, p-value=0.009901 |
|---|---|---|---|---|
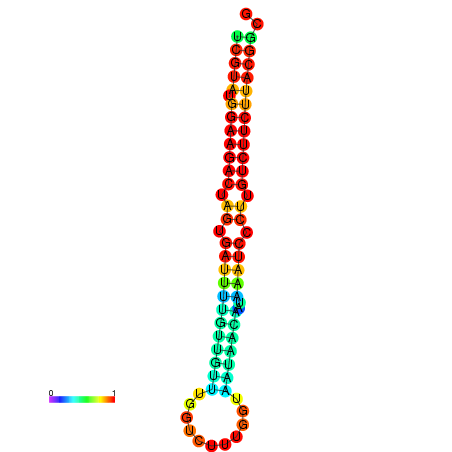 |
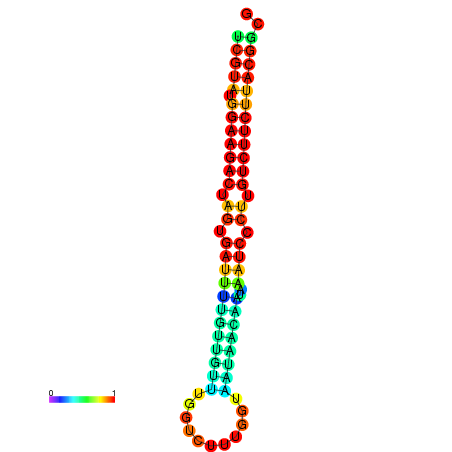 |
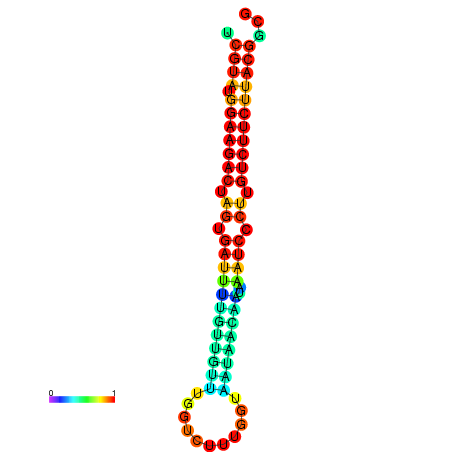 |
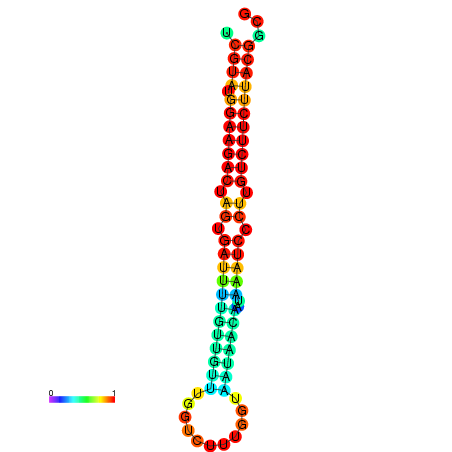 |
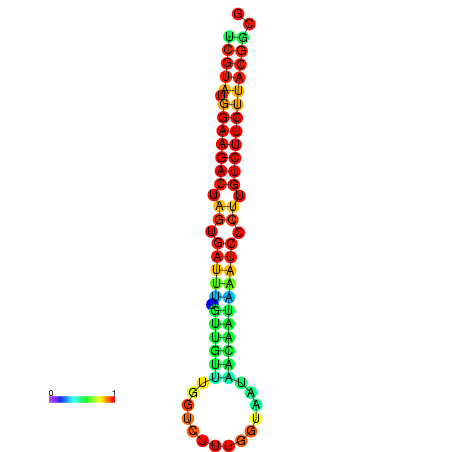 |
| dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|---|
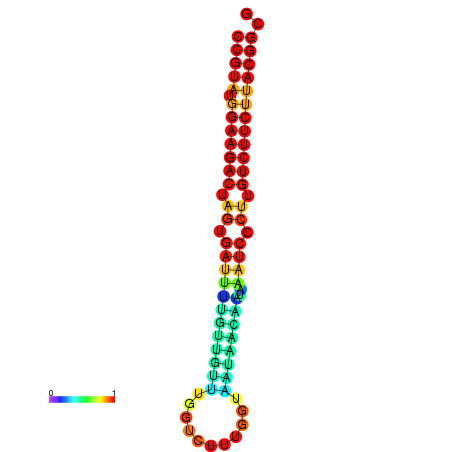
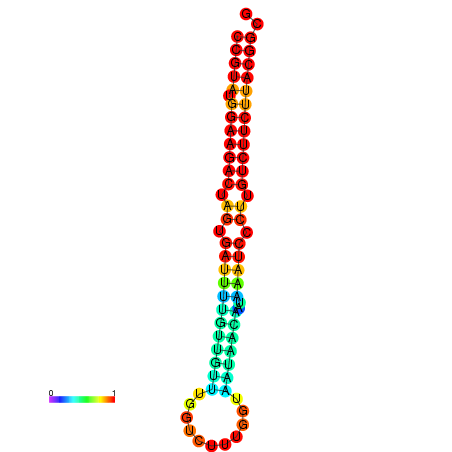 |
 |
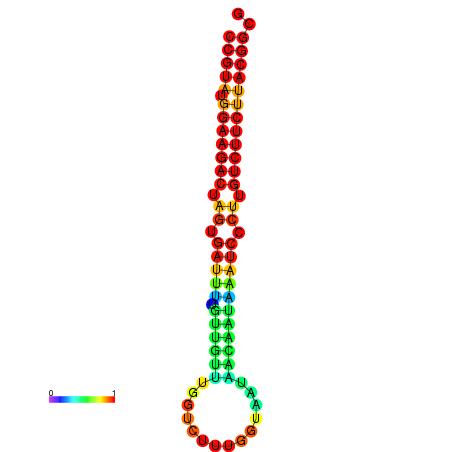 |
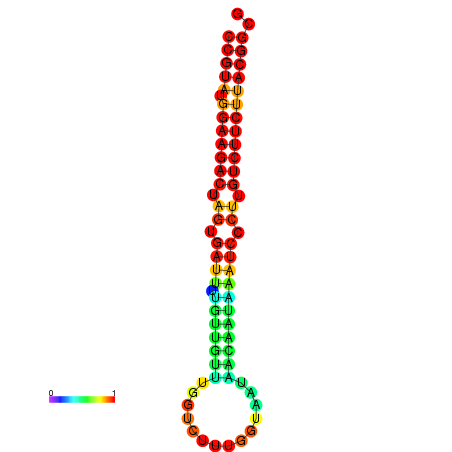 |
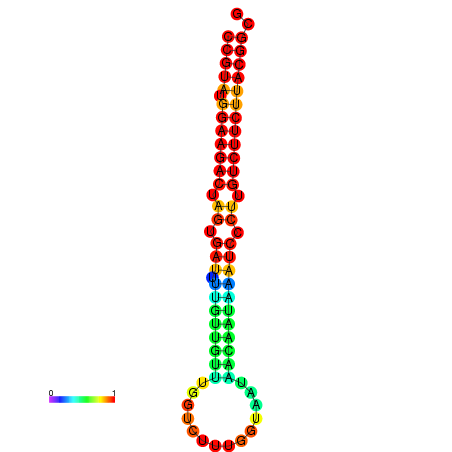 |
| dG=-23.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|---|
 |
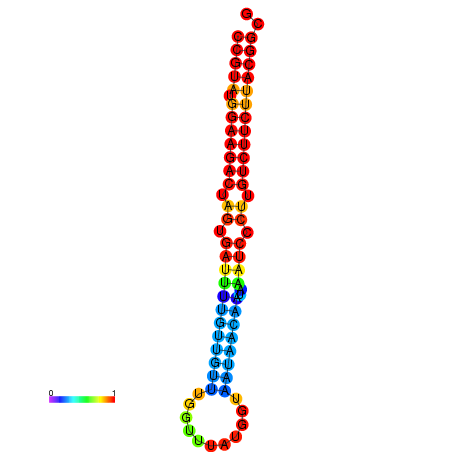 |
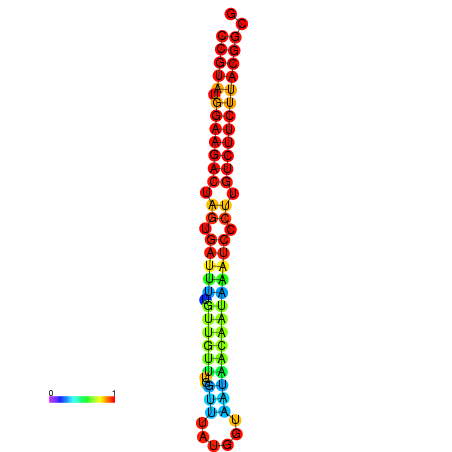 |
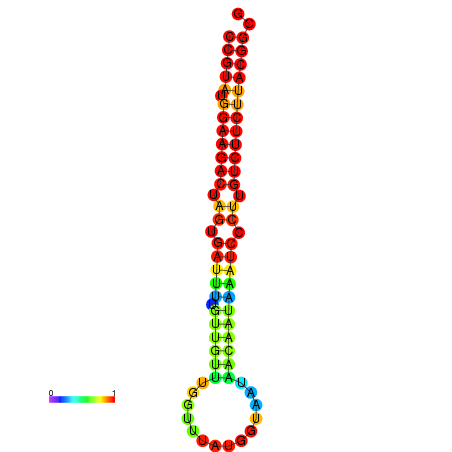 |
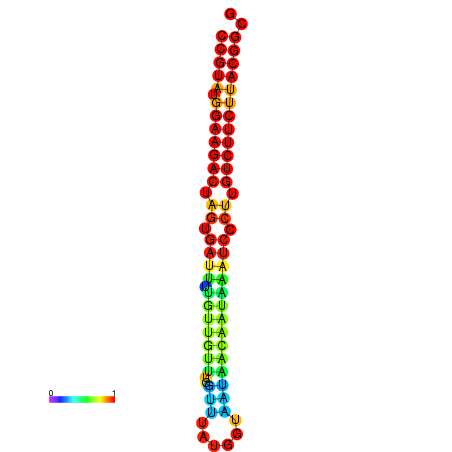 |
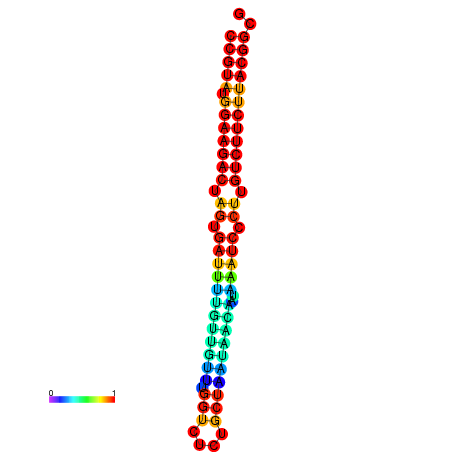
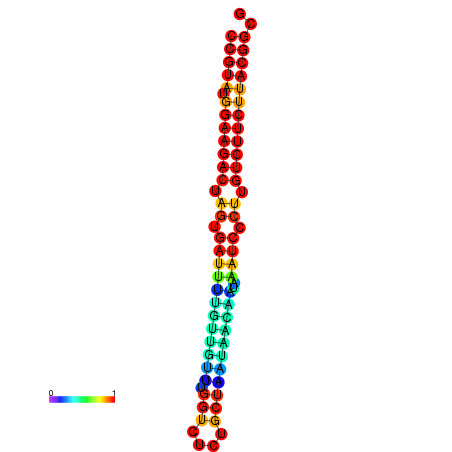
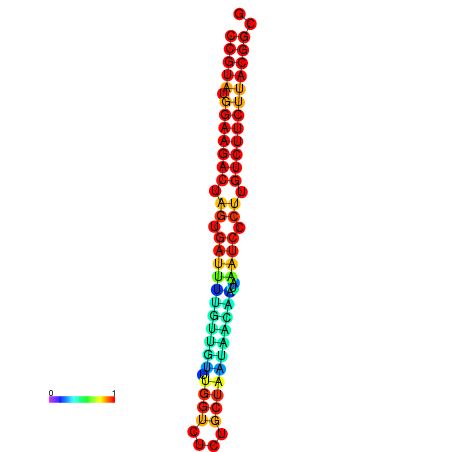
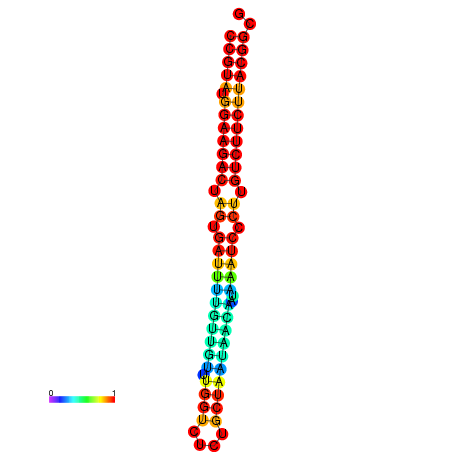
| dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
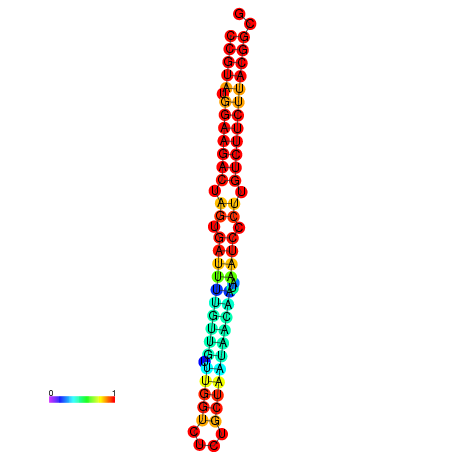 |
| dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 |
|---|---|---|---|---|
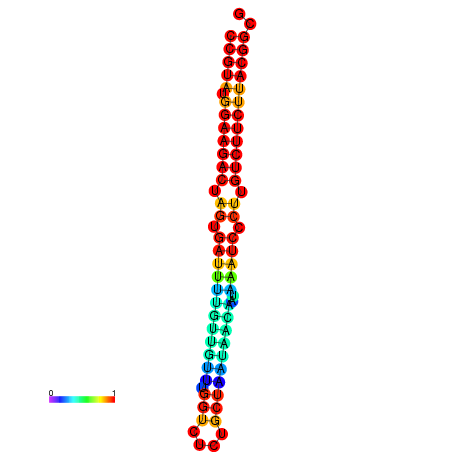 |
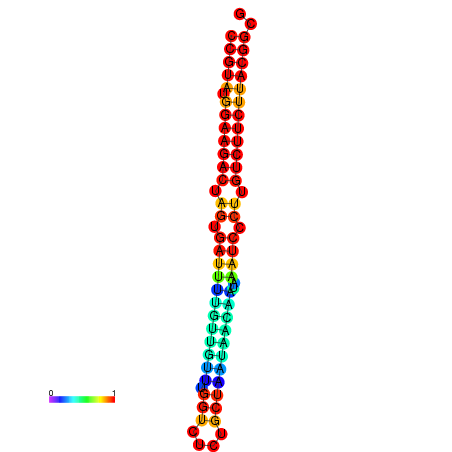 |
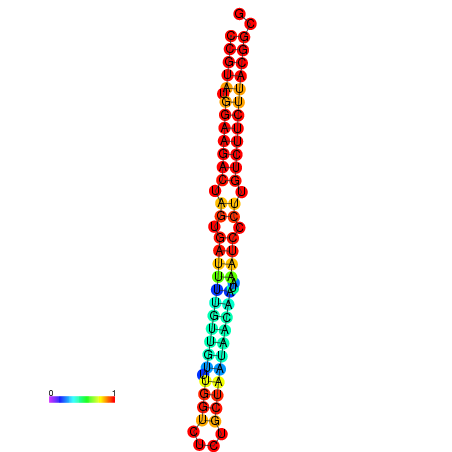 |
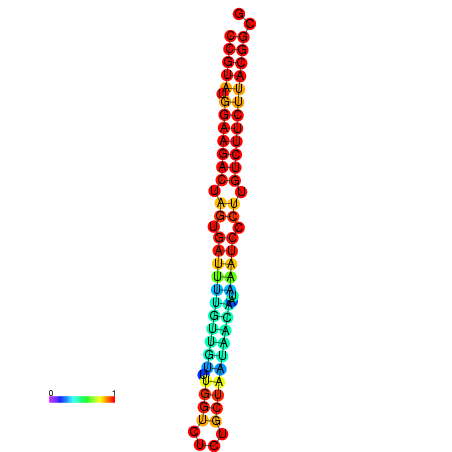 |
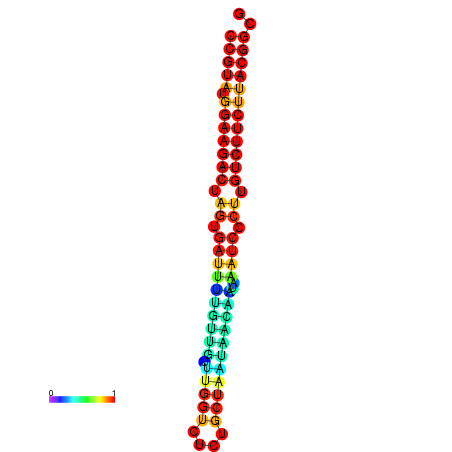 |
| dG=-21.2, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-20.8, p-value=0.009901 |
|---|---|---|---|---|
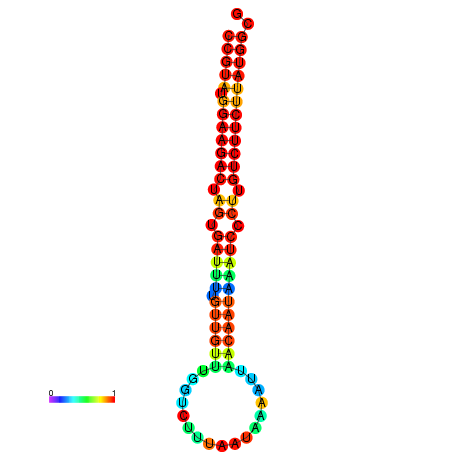 |
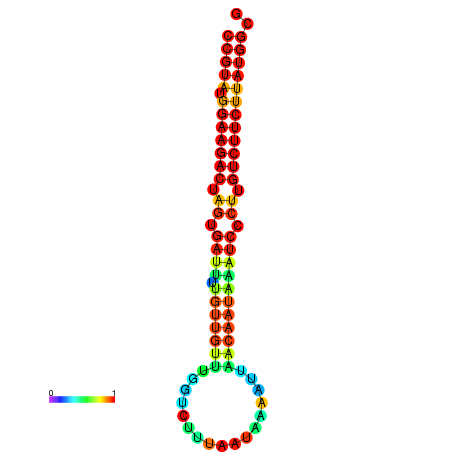 |
 |
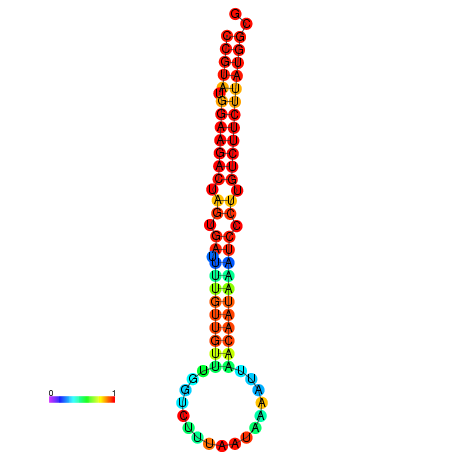 |
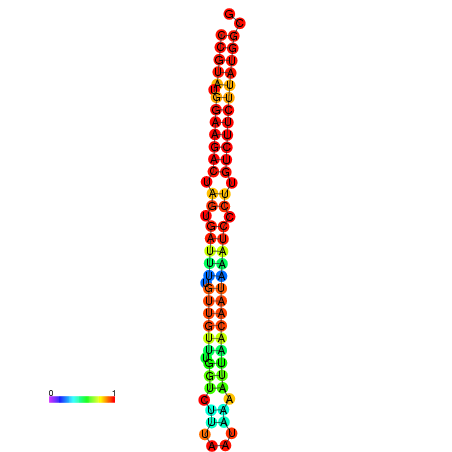 |
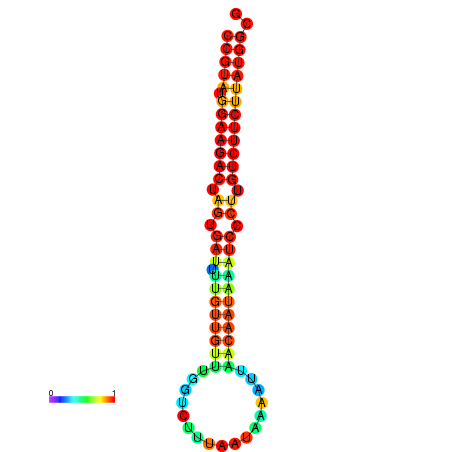
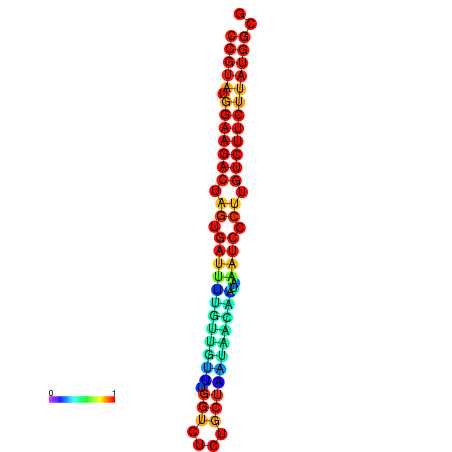
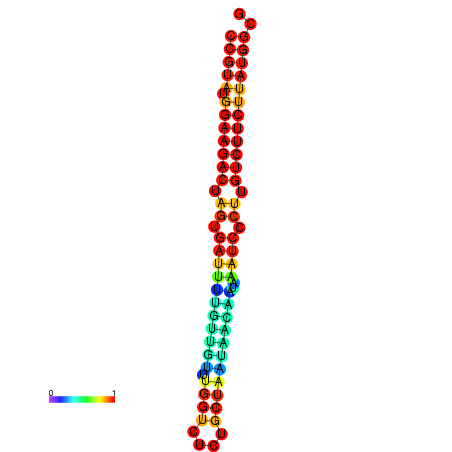
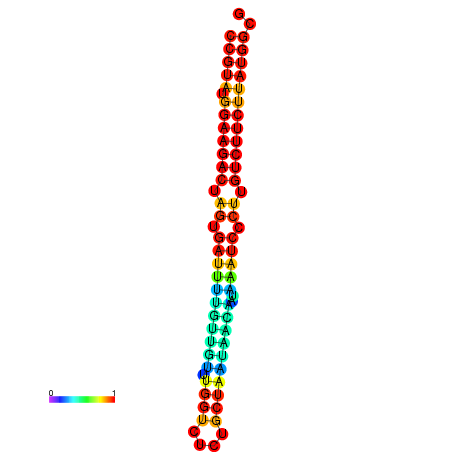
| dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
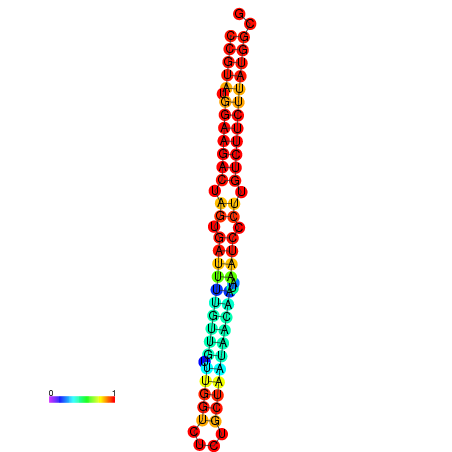 |
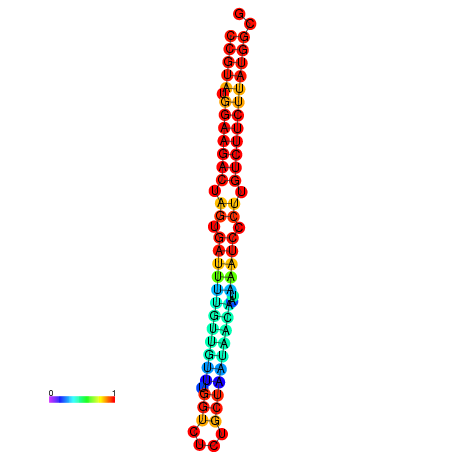
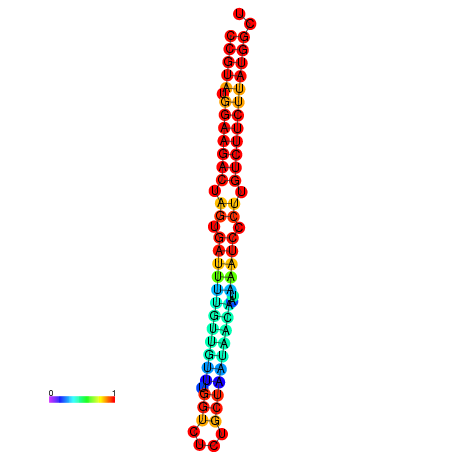
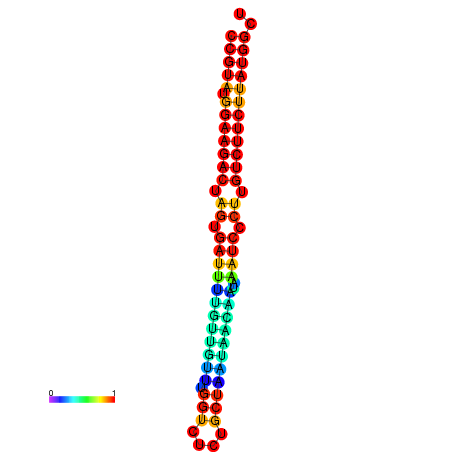
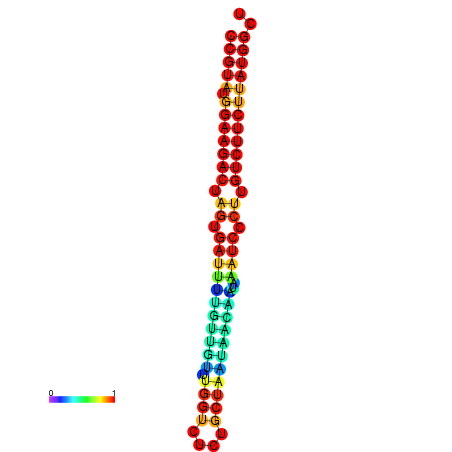
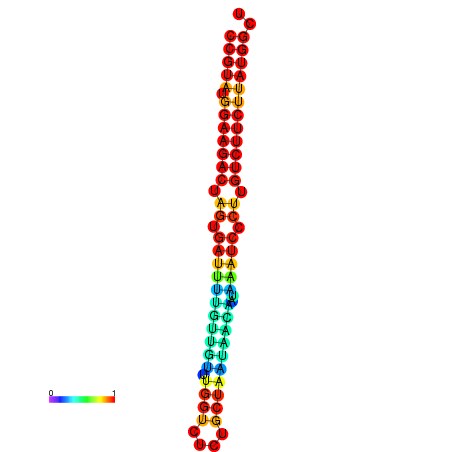
| dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 | dG=-23.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
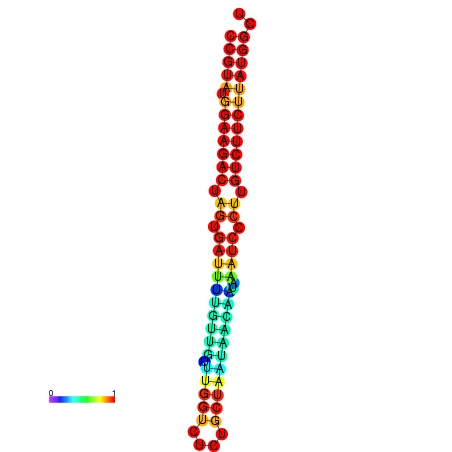 |
| dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-20.1, p-value=0.009901 | dG=-19.8, p-value=0.009901 |
|---|---|---|---|---|
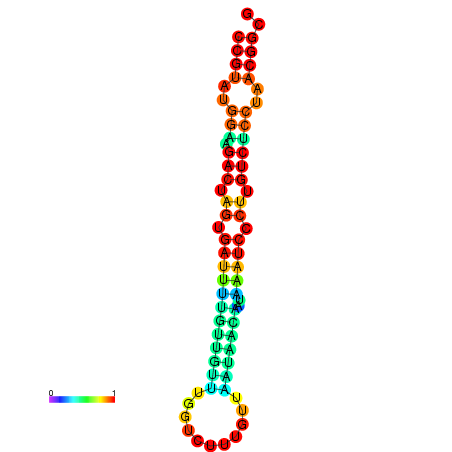 |
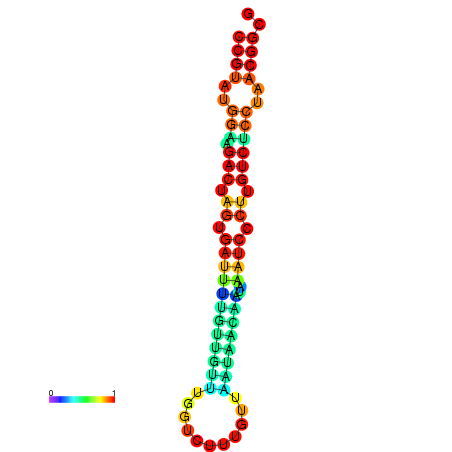 |
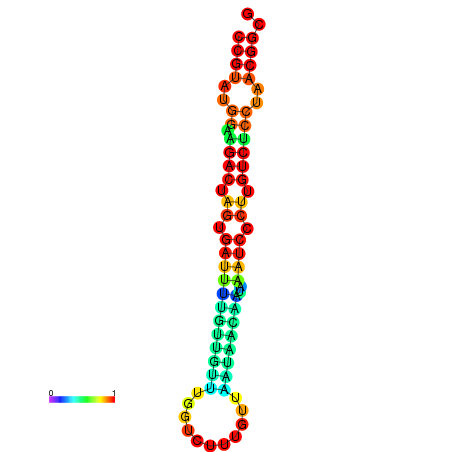 |
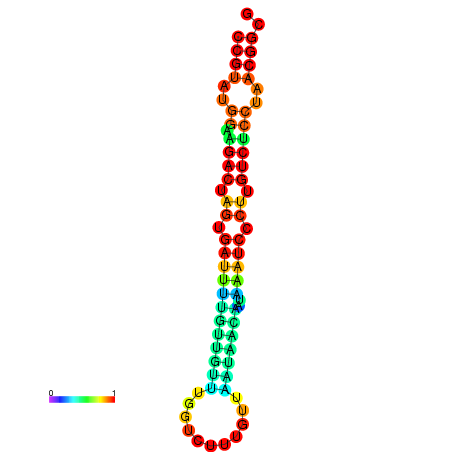 |
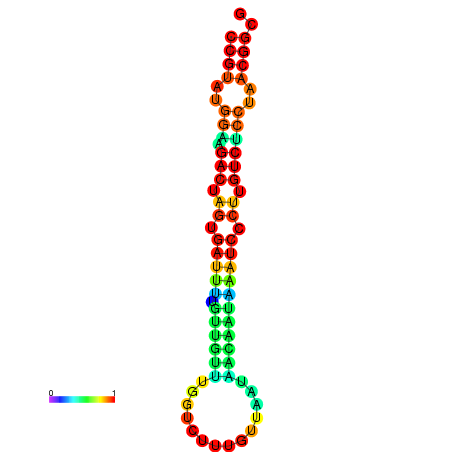 |
Generated: 03/07/2013 at 07:01 PM