
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548206-15548315 - | AAA-----TTGAAAATTGTACAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTTGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGGCAATCGATCTACGT |
| droSim1 | chr2R:14253641-14253750 - | AAA-----TTGAAAATTGTACAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTCGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGGCAATCGATCTACGT |
| droSec1 | super_1:13062939-13063048 - | AAA-----TTGAAAATTGTACAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTCGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGGCAATCGATCTACGT |
| droYak2 | chr2R:13624474-13624583 + | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTCGAAACTCT----CTCAATTTATATCACAGTGGCTGTTCTTTTTTATTTGGCAATCGATCTACAT |
| droEre2 | scaffold_4845:9748460-9748569 - | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTGGCTGATGATGTAGTTCGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCTGCGT |
| droAna3 | scaffold_13266:9732107-9732212 + | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTCGCTGCTGATGTAGTT----ATTCG----CATCATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCTACGT |
| dp4 | chr3:12676789-12676898 - | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTTGCTGCTGATGTAGTTCAAGTTTTG----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCACCGA |
| droPer1 | super_2:8906774-8906883 + | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTTGCTGCTGATGTAGTTCAAGTTTCG----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCAACGA |
| droWil1 | scaffold_180701:1650985-1651090 - | CAT-----TT----TTTGTTTAAAAAGAAGGGAACGGTTGCTGTTGATGTAGTATACCATTTTGTTTAACAA--TATATCACAGTGGCTGTTCTTTTTTGTTTAACAATCAATCAAC-- |
| droVir3 | scaffold_12875:4112942-4113050 + | CGC-----TCGTCAATTGTTTAAAAGGAAGAGAACGTTCGCTGTTGATGTAGT-CGACCTTTG----TACTCTTTATATCACAGTGGCTGTTCTTTTTTGTTTAGCAATTTCTCAACTT |
| droMoj3 | scaffold_6496:20935943-20936057 - | AATCGTGGCCGTAAATTGCTTAAAGAGAAGAGAACGTTCGCTGTTGATGTAGTCGAATTCATT----TAATCTTTATATCACAGTGGCTGTTCTTTTTTGTTTAACAATTTTTCAACGT |
| droGri2 | scaffold_15112:2107905-2108014 - | A-C-----TCATAAATTGTTTAAAAGGAAGAGAACGCTCGCTGCTGATGTAGTTCATGATTCA----AATATTTTATATCACAGTGGCTGTTCTTTTTTGTTTATCAATGATTCAACTT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548206-15548315 - | AAA-----TTGAAAATTGTACAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTTGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGGCAATCGATCTACGT |
| droSim1 | chr2R:14253641-14253750 - | AAA-----TTGAAAATTGTACAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTCGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGGCAATCGATCTACGT |
| droSec1 | super_1:13062939-13063048 - | AAA-----TTGAAAATTGTACAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTCGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGGCAATCGATCTACGT |
| droYak2 | chr2R:13624474-13624583 + | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTTGCTGATGATGTAGTTCGAAACTCT----CTCAATTTATATCACAGTGGCTGTTCTTTTTTATTTGGCAATCGATCTACAT |
| droEre2 | scaffold_4845:9748460-9748569 - | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTGGCTGATGATGTAGTTCGAAACTCT----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCTGCGT |
| droAna3 | scaffold_13266:9732107-9732212 + | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTCGCTGCTGATGTAGTT----ATTCG----CATCATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCTACGT |
| dp4 | chr3:12676789-12676898 - | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTTGCTGCTGATGTAGTTCAAGTTTTG----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCACCGA |
| droPer1 | super_2:8906774-8906883 + | AAA-----TTGAAAATTGTTCAAAAAGAAGGGAACGGTTGCTGCTGATGTAGTTCAAGTTTCG----CACAATTTATATCACAGTGGCTGTTCTTTTTTGTTTGCCAATCGATCAACGA |
| droWil1 | scaffold_180701:1650985-1651090 - | CAT-----TT----TTTGTTTAAAAAGAAGGGAACGGTTGCTGTTGATGTAGTATACCATTTTGTTTAACAA--TATATCACAGTGGCTGTTCTTTTTTGTTTAACAATCAATCAAC-- |
| droVir3 | scaffold_12875:4112942-4113050 + | CGC-----TCGTCAATTGTTTAAAAGGAAGAGAACGTTCGCTGTTGATGTAGT-CGACCTTTG----TACTCTTTATATCACAGTGGCTGTTCTTTTTTGTTTAGCAATTTCTCAACTT |
| droMoj3 | scaffold_6496:20935943-20936057 - | AATCGTGGCCGTAAATTGCTTAAAGAGAAGAGAACGTTCGCTGTTGATGTAGTCGAATTCATT----TAATCTTTATATCACAGTGGCTGTTCTTTTTTGTTTAACAATTTTTCAACGT |
| droGri2 | scaffold_15112:2107905-2108014 - | A-C-----TCATAAATTGTTTAAAAGGAAGAGAACGCTCGCTGCTGATGTAGTTCATGATTCA----AATATTTTATATCACAGTGGCTGTTCTTTTTTGTTTATCAATGATTCAACTT |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-29.5, p-value=0.009901 | dG=-29.1, p-value=0.009901 | dG=-28.6, p-value=0.009901 |
|---|---|---|
 |
 |
 |
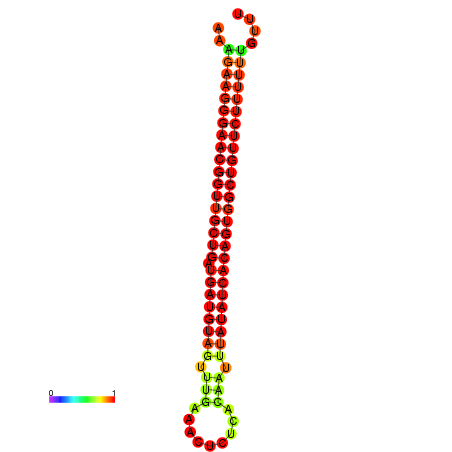
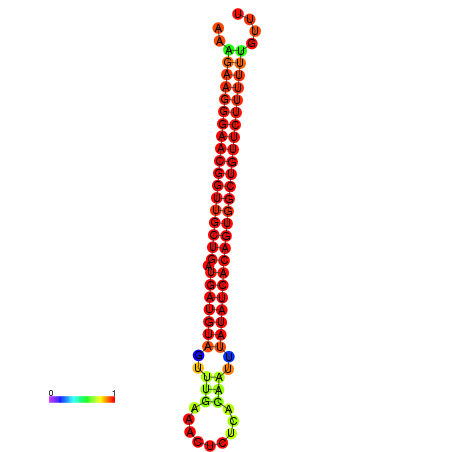
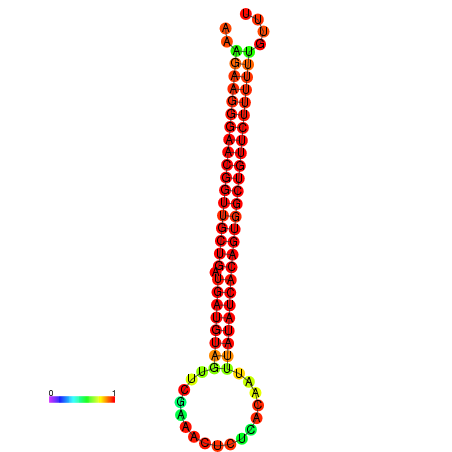
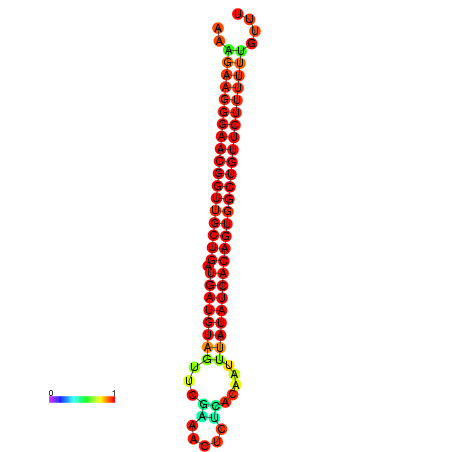
| dG=-26.9, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.2, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
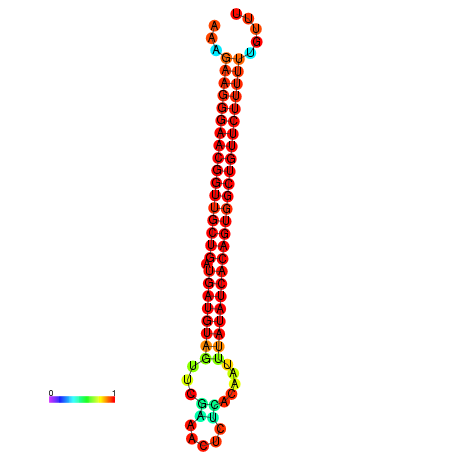 |
| dG=-26.9, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.2, p-value=0.009901 |
|---|---|---|---|
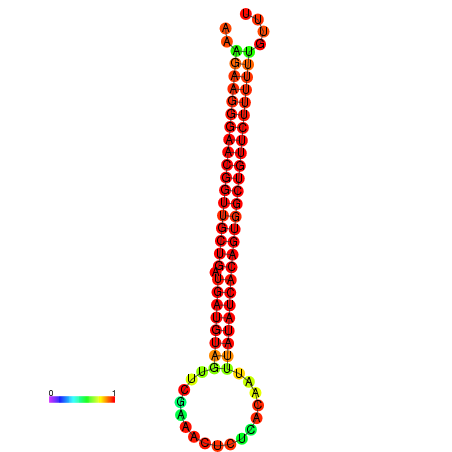 |
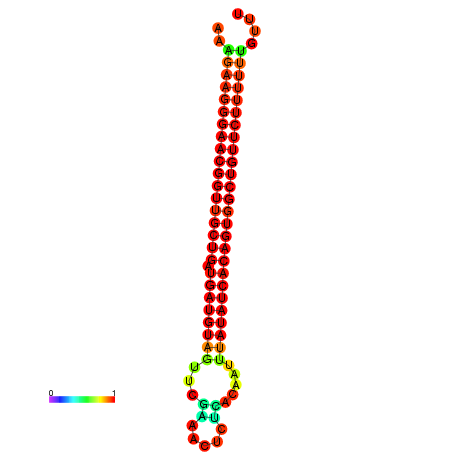 |
 |
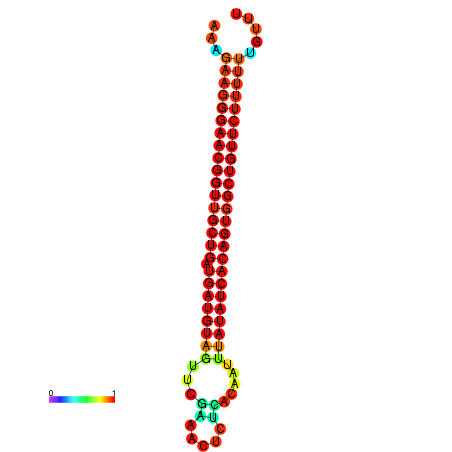 |
| dG=-27.8, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-26.9, p-value=0.009901 |
|---|---|---|
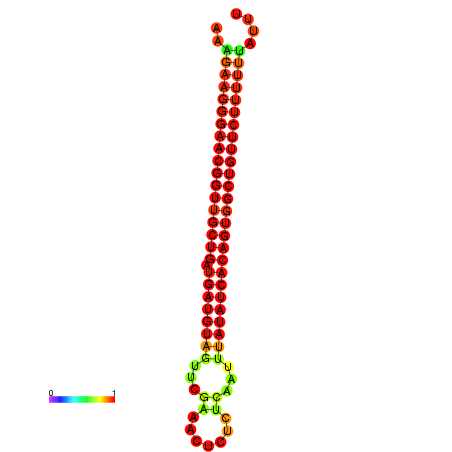 |
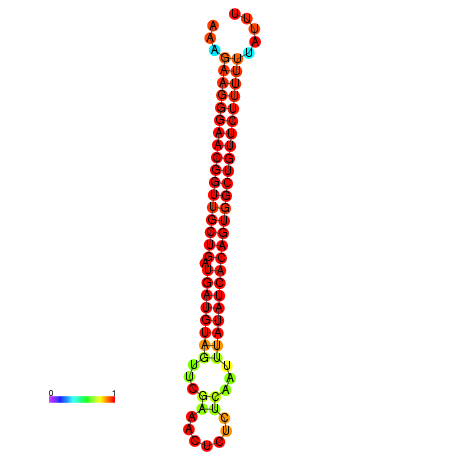 |
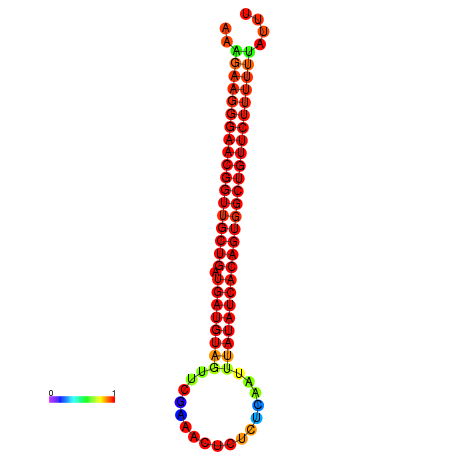 |
| dG=-27.4, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|---|---|
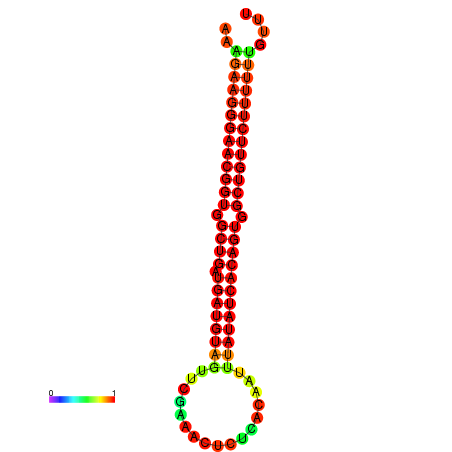 |
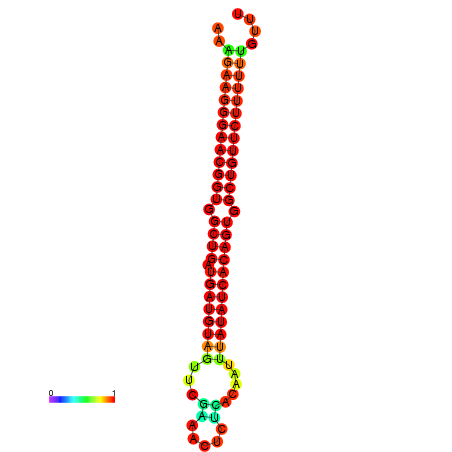 |
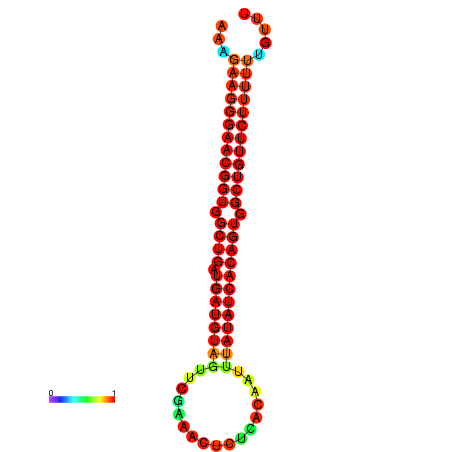 |
 |
| dG=-29.9, p-value=0.009901 | dG=-29.5, p-value=0.009901 |
|---|---|
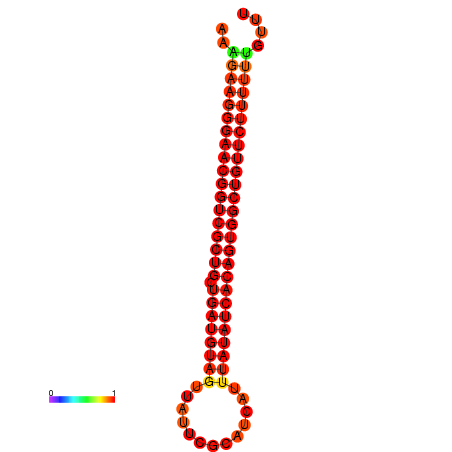 |
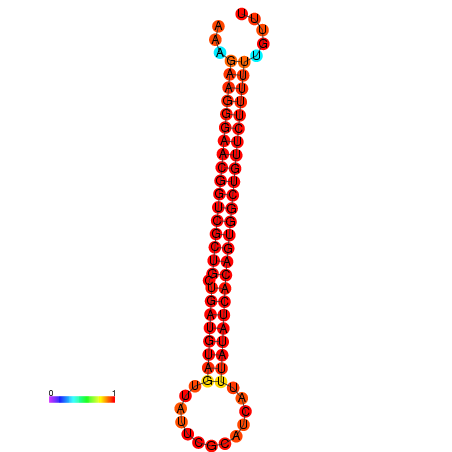 |
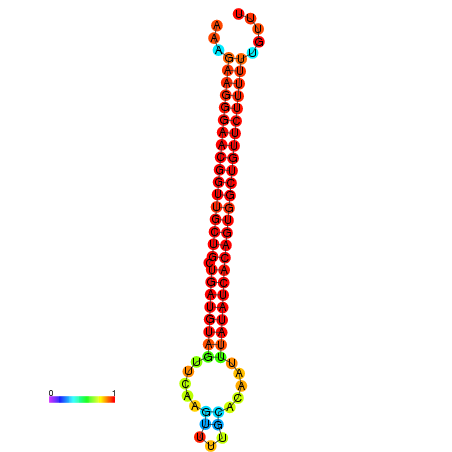
| dG=-27.0, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
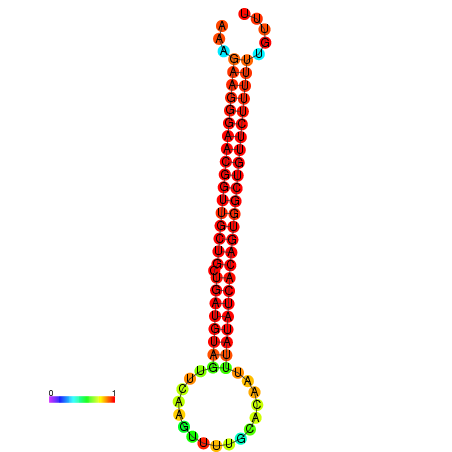 |
 |
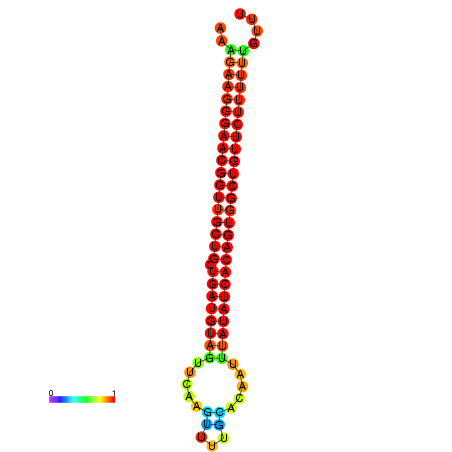
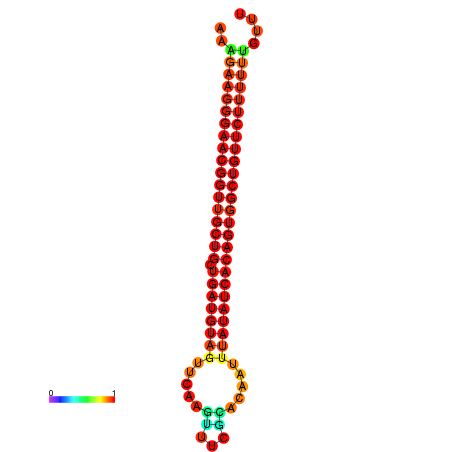
| dG=-27.0, p-value=0.009901 | dG=-26.9, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
 |
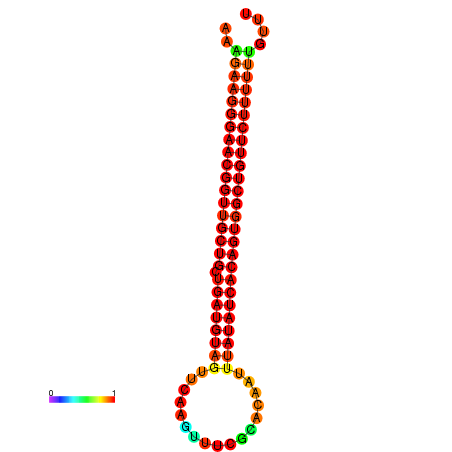 |
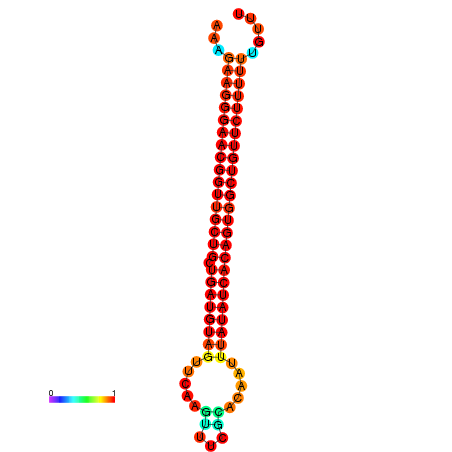 |
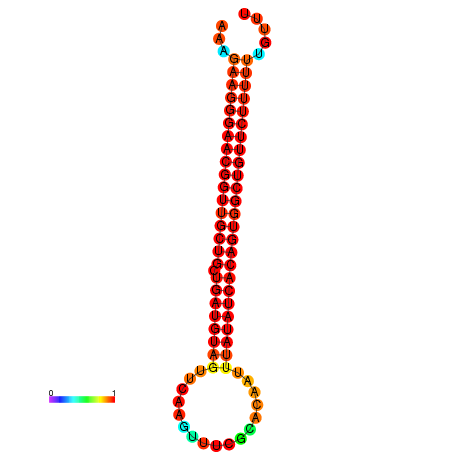 |
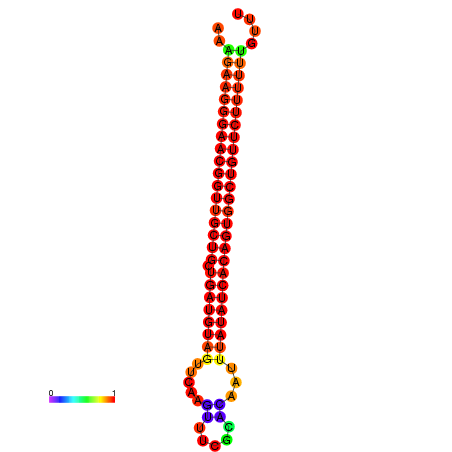 |
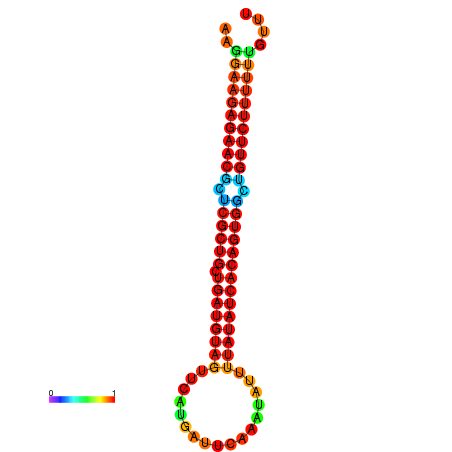
| dG=-26.3, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
 |
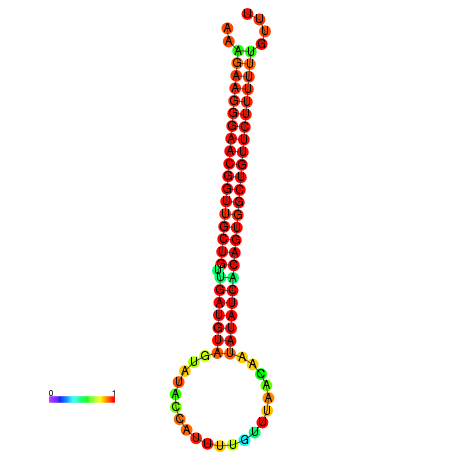 |
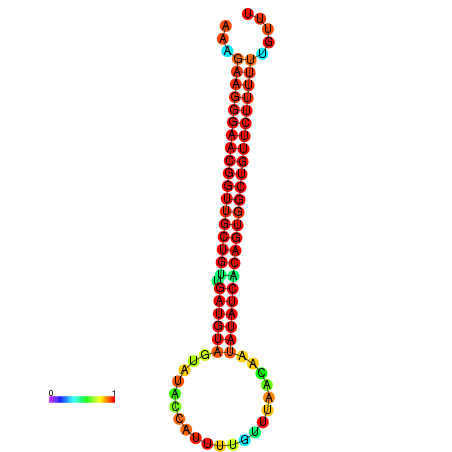 |
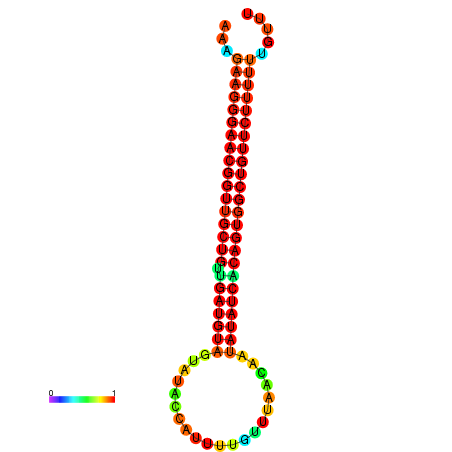 |
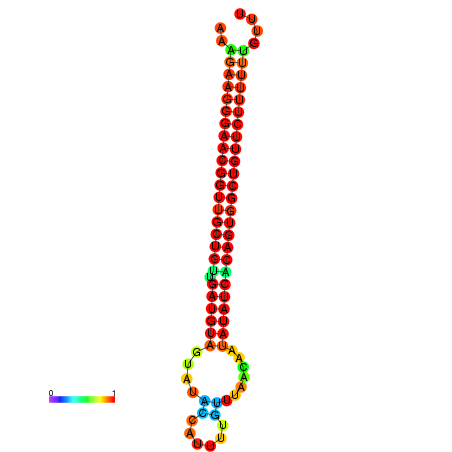 |
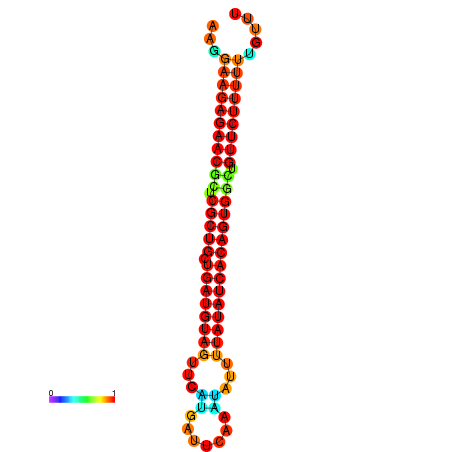
| dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
 |
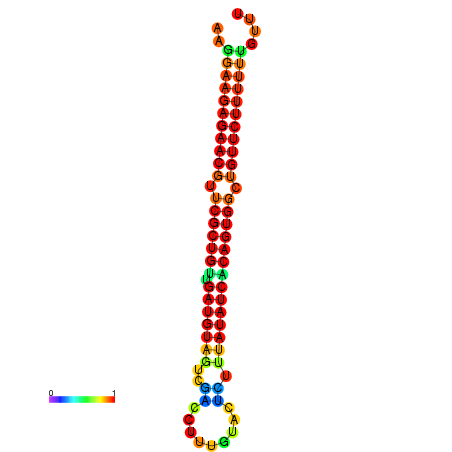 |
 |
 |
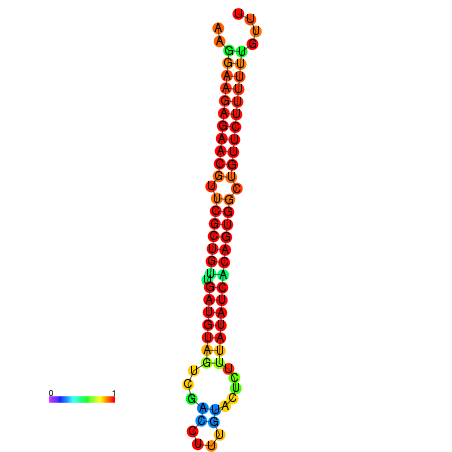 |
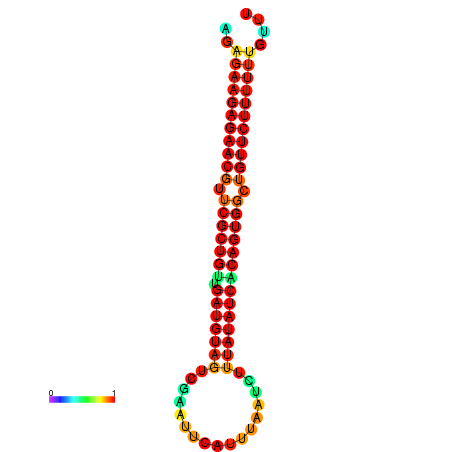
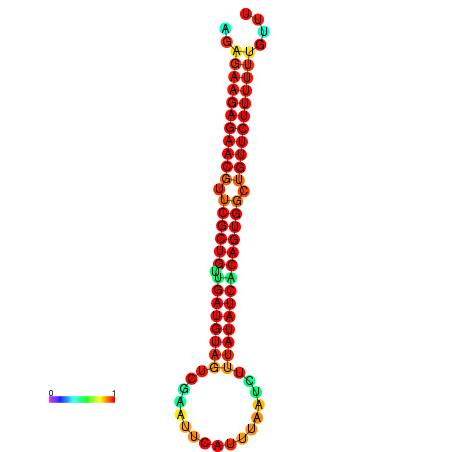
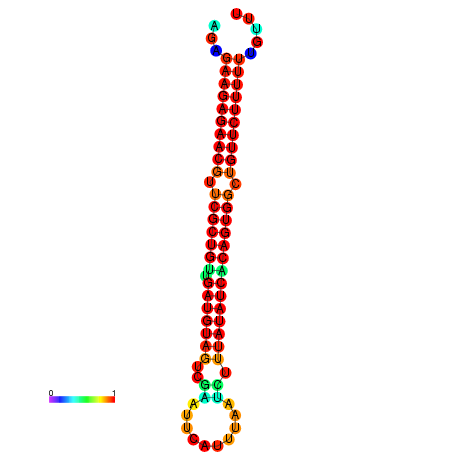
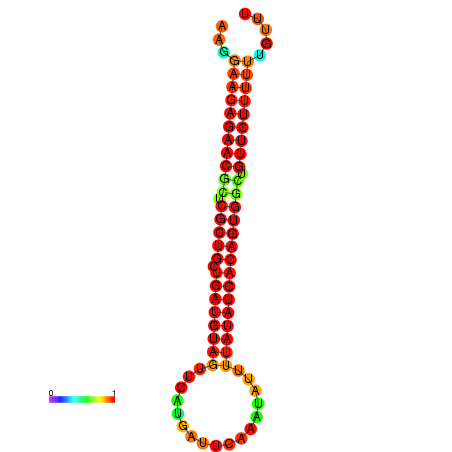
| dG=-24.0, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.6, p-value=0.009901 |
|---|---|---|---|---|
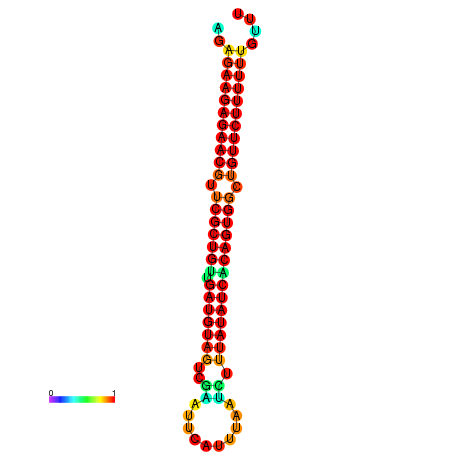 |
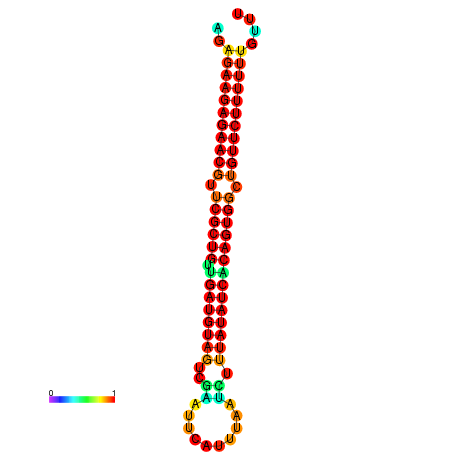 |
 |
 |
 |
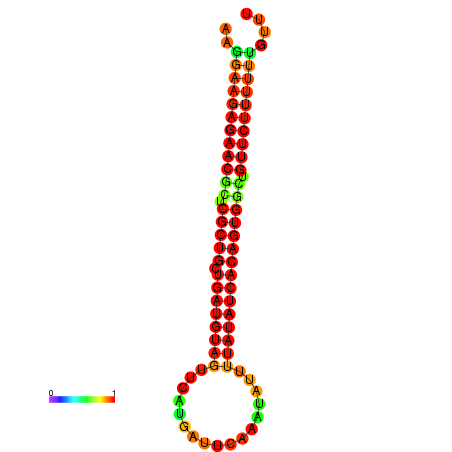
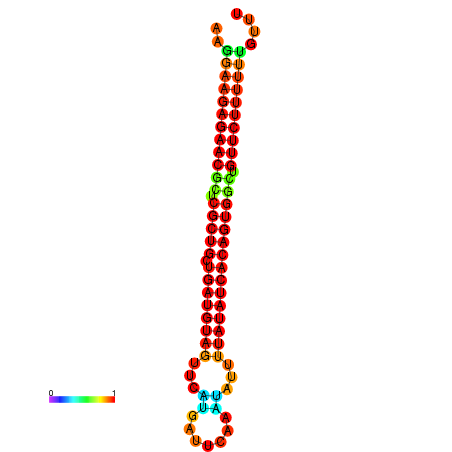
| dG=-24.3, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.9, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 07:00 PM