| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548359-15548462 - | TAACCA---------------------TGCCG--TTTTTAACCCAAGGGAACTTCTGCTGCTGATATATTAT-----------TGA-------------AAAA----CTACTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droSim1 | chr2R:14253794-14253897 - | TAACCA---------------------TGCCG--TTTTTAACCCCAGGGAACTTCTGCTGCTGATATATTAT-----------TGA-------------AAAA----CTCCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droSec1 | super_1:13063092-13063195 - | TAACCA---------------------TGCCG--TTTTTAACCCCAGGGAACTTCTGCTGCTGATATATTAT-----------TGA-------------AAAA----CTCCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droYak2 | chr2R:13624327-13624430 + | TAACCA---------------------TACCG--TTTTTAACCGCAGGGAACCTCTGCTGCTGATATATTAT-----------TGA-------------GAAA----CTTCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droEre2 | scaffold_4845:9748613-9748715 - | AACCCA---------------------TACCG--TTTTTAACCCCAGGGAACCTCTGCTGCTGATATATTAT-----------TGA--------------AAA----CTCCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAG |
| droAna3 | scaffold_13266:9731948-9732061 + | ATATTA---------------------TTCCG--GTGTTAGCCCCAGGGAACTGTTGCTGCTGATATATTATTCTCCCAAAAA-AA-------------AAAT----CACCTATATCACAGTGGCTGTTCTTTTTGGCTGCACCGACAAG-TTCAC |
| dp4 | chr3:12676931-12677045 - | TCCCCG---------------------TGCCGTGTCCCCAACCGCAGGGAACCGCTGCTGCTGATATATTATCCACCCATCTTTGA---------------CA----TTTCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAG-TCCAA |
| droPer1 | super_2:8906627-8906741 + | TCCCCG---------------------TGCCGTGTCCCCAACCGCAGGGAACCGCTGCTGCTGATATATTATCCACCCATCTTTGA---------------CA----TTTCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAG-TCCAA |
| droWil1 | scaffold_180701:1651131-1651259 - | TTTTTA---------------------AAGTTTTTGTTTAACCGCAGGGAACTGTCGCTGCTGATATAT-----ATATATTATTGTTATATATAATTTAAAAATGATTTTCTATATCACAGTGGCTGTTCTTTTTGGTTTTACGATCATT-TTCAA |
| droVir3 | scaffold_12875:4112748-4112878 + | TAAATATTTGCTGACAAATTTCTTATGTGCTA--TGTTTAACCGCAGGGAACTGCTGCTGCTGATATACTGTATAGATTATATCGA----------------------TTCCATATCACAGTGGCTGTTCTTTTTGGTTGCATCGCAATC-CACAA |
| droMoj3 | scaffold_6496:20936121-20936236 - | TATCCA------GAGA-----------GATTG--CGTTTAACCGCAGGGAACTGCTGCTGCTGATATACTATA----TGTATTTGT---------------CA--GTACTCTATATCACAGTGGCTGTTCTTTTTGGTTGCATCGCAAATAGCCAA |
| droGri2 | scaffold_15112:2108071-2108181 - | TTCACC-------------------AATGCTA--TGTTTAACCACAGGGAACTGCTGCTGCTGATATAC-----AACCATAT-TTA-------------TACA----CGACTATATCACAGTGGCTGTTCTTTTTGGTTGCATCGCCATT-TGCAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548359-15548462 - | TAACCA---------------------TGCCG--TTTTTAACCCAAGGGAACTTCTGCTGCTGATATATTAT-----------TGA-------------AAAA----CTACTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droSim1 | chr2R:14253794-14253897 - | TAACCA---------------------TGCCG--TTTTTAACCCCAGGGAACTTCTGCTGCTGATATATTAT-----------TGA-------------AAAA----CTCCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droSec1 | super_1:13063092-13063195 - | TAACCA---------------------TGCCG--TTTTTAACCCCAGGGAACTTCTGCTGCTGATATATTAT-----------TGA-------------AAAA----CTCCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droYak2 | chr2R:13624327-13624430 + | TAACCA---------------------TACCG--TTTTTAACCGCAGGGAACCTCTGCTGCTGATATATTAT-----------TGA-------------GAAA----CTTCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAA |
| droEre2 | scaffold_4845:9748613-9748715 - | AACCCA---------------------TACCG--TTTTTAACCCCAGGGAACCTCTGCTGCTGATATATTAT-----------TGA--------------AAA----CTCCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAT-TCCAG |
| droAna3 | scaffold_13266:9731948-9732061 + | ATATTA---------------------TTCCG--GTGTTAGCCCCAGGGAACTGTTGCTGCTGATATATTATTCTCCCAAAAA-AA-------------AAAT----CACCTATATCACAGTGGCTGTTCTTTTTGGCTGCACCGACAAG-TTCAC |
| dp4 | chr3:12676931-12677045 - | TCCCCG---------------------TGCCGTGTCCCCAACCGCAGGGAACCGCTGCTGCTGATATATTATCCACCCATCTTTGA---------------CA----TTTCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAG-TCCAA |
| droPer1 | super_2:8906627-8906741 + | TCCCCG---------------------TGCCGTGTCCCCAACCGCAGGGAACCGCTGCTGCTGATATATTATCCACCCATCTTTGA---------------CA----TTTCTATATCACAGTGGCTGTTCTTTTTGGTTGCACGGCCAAG-TCCAA |
| droWil1 | scaffold_180701:1651131-1651259 - | TTTTTA---------------------AAGTTTTTGTTTAACCGCAGGGAACTGTCGCTGCTGATATAT-----ATATATTATTGTTATATATAATTTAAAAATGATTTTCTATATCACAGTGGCTGTTCTTTTTGGTTTTACGATCATT-TTCAA |
| droVir3 | scaffold_12875:4112748-4112878 + | TAAATATTTGCTGACAAATTTCTTATGTGCTA--TGTTTAACCGCAGGGAACTGCTGCTGCTGATATACTGTATAGATTATATCGA----------------------TTCCATATCACAGTGGCTGTTCTTTTTGGTTGCATCGCAATC-CACAA |
| droMoj3 | scaffold_6496:20936121-20936236 - | TATCCA------GAGA-----------GATTG--CGTTTAACCGCAGGGAACTGCTGCTGCTGATATACTATA----TGTATTTGT---------------CA--GTACTCTATATCACAGTGGCTGTTCTTTTTGGTTGCATCGCAAATAGCCAA |
| droGri2 | scaffold_15112:2108071-2108181 - | TTCACC-------------------AATGCTA--TGTTTAACCACAGGGAACTGCTGCTGCTGATATAC-----AACCATAT-TTA-------------TACA----CGACTATATCACAGTGGCTGTTCTTTTTGGTTGCATCGCCATT-TGCAA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-23.4, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.2, p-value=0.009901 | dG=-22.6, p-value=0.009901 | dG=-22.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
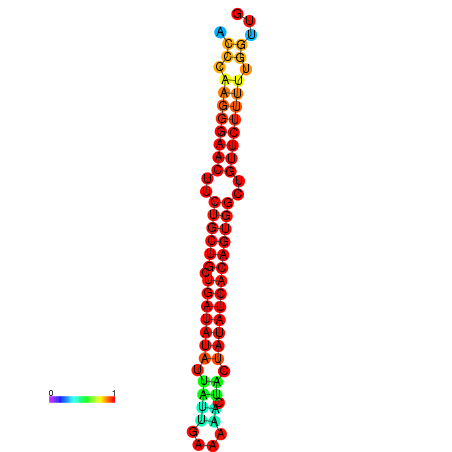
| dG=-21.5, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.7, p-value=0.009901 |
|---|---|---|
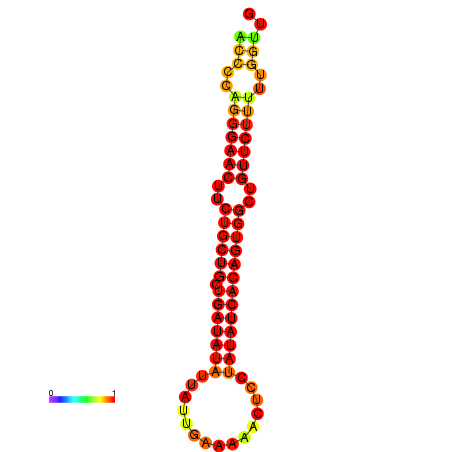 |
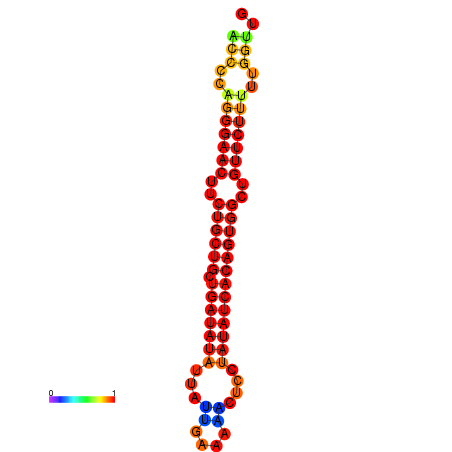 |
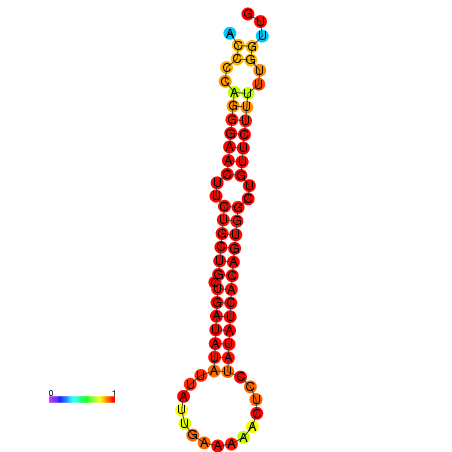 |
| dG=-21.5, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.7, p-value=0.009901 |
|---|---|---|
 |
 |
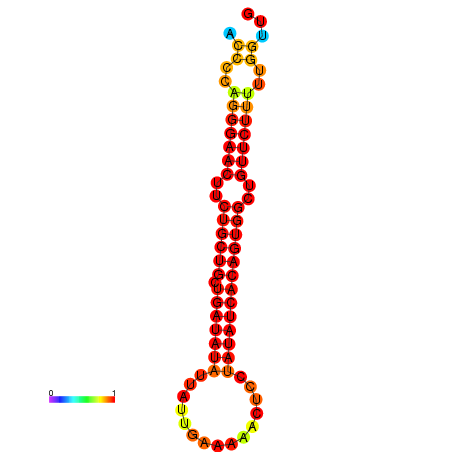 |
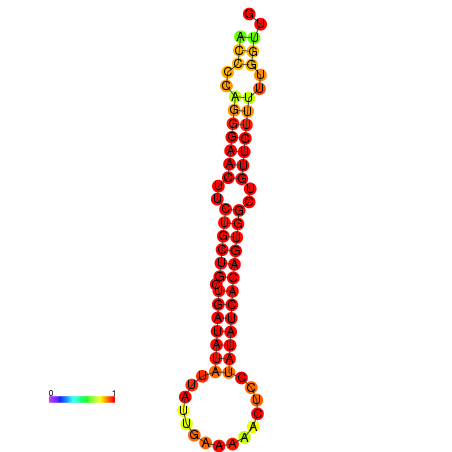
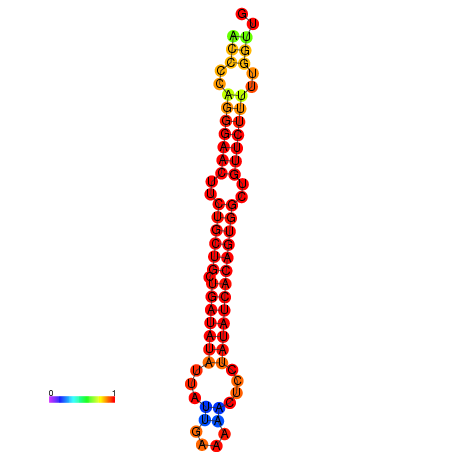
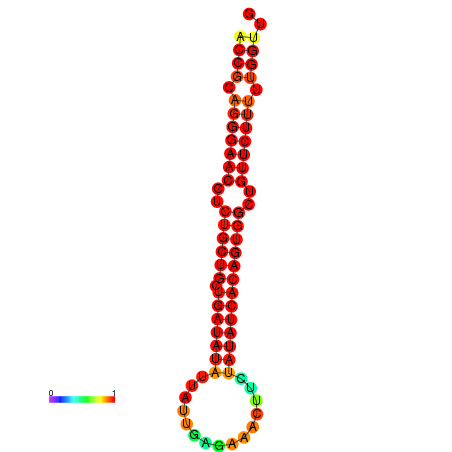
| dG=-22.2, p-value=0.009901 | dG=-22.2, p-value=0.009901 | dG=-21.8, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
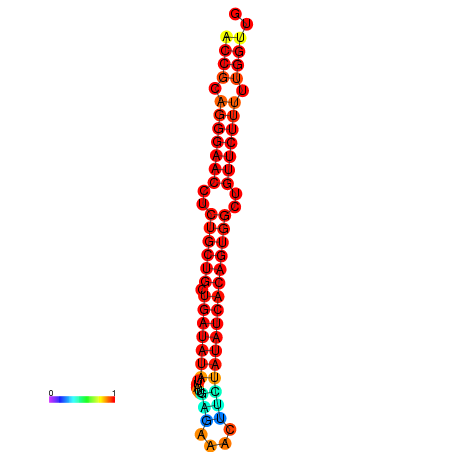
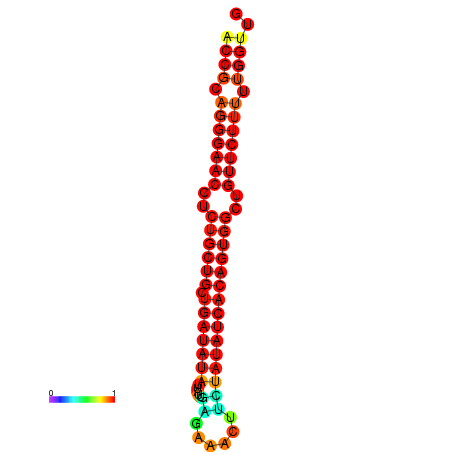
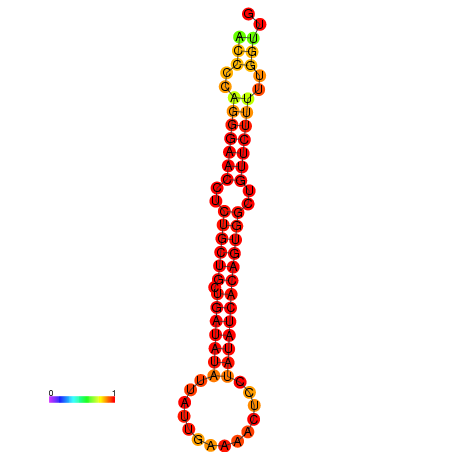
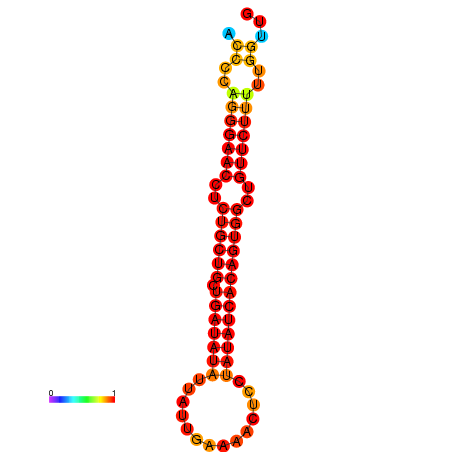
| dG=-20.6, p-value=0.009901 | dG=-19.8, p-value=0.009901 |
|---|---|
 |
 |
| dG=-23.4, p-value=0.009901 |
|---|
 |
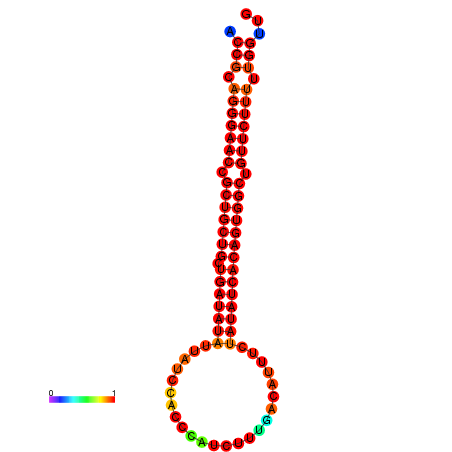
| dG=-25.8, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
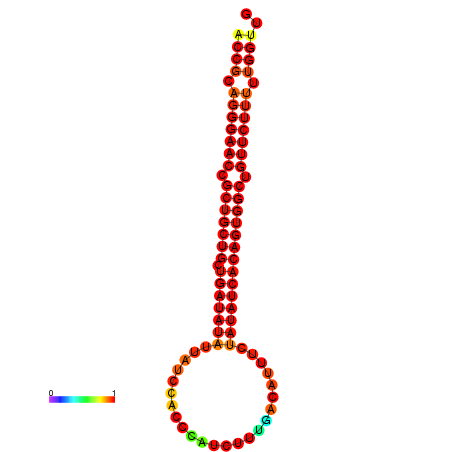 |
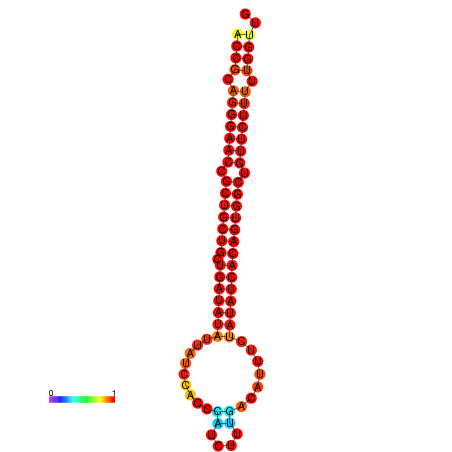 |
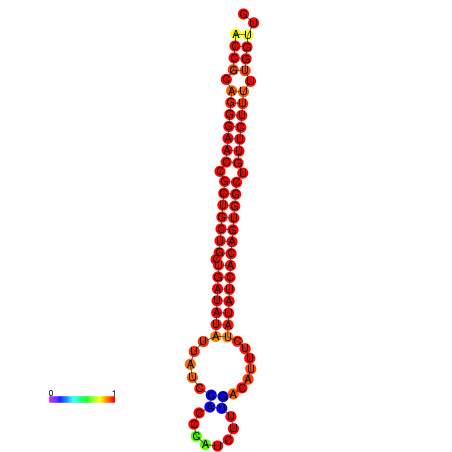 |
 |
 |
| dG=-25.8, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-25.0, p-value=0.009901 | dG=-24.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
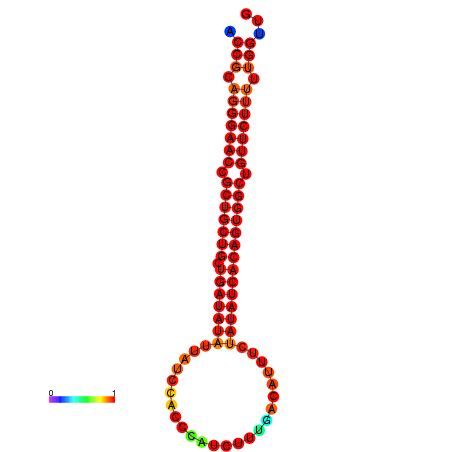 |
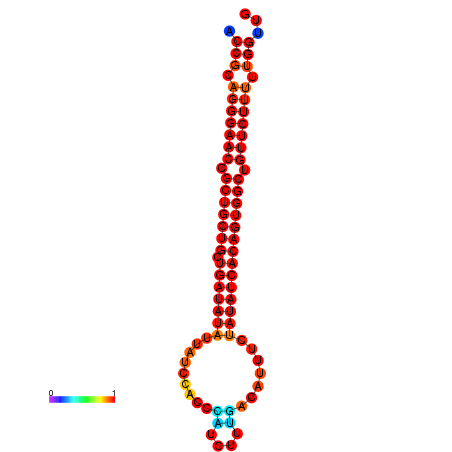 |
| dG=-29.9, p-value=0.009901 | dG=-29.3, p-value=0.009901 | dG=-29.2, p-value=0.009901 | dG=-29.1, p-value=0.009901 | dG=-29.1, p-value=0.009901 |
|---|---|---|---|---|
 |
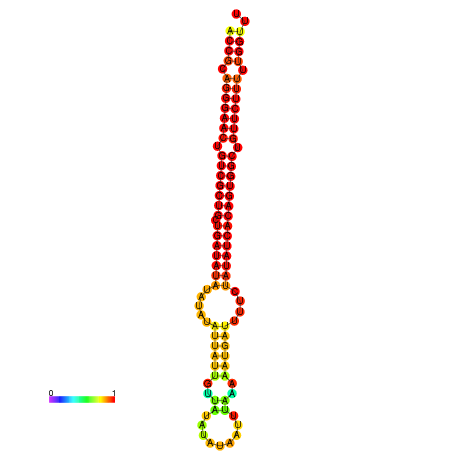 |
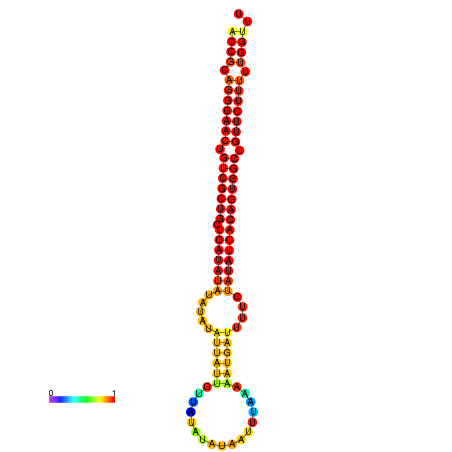 |
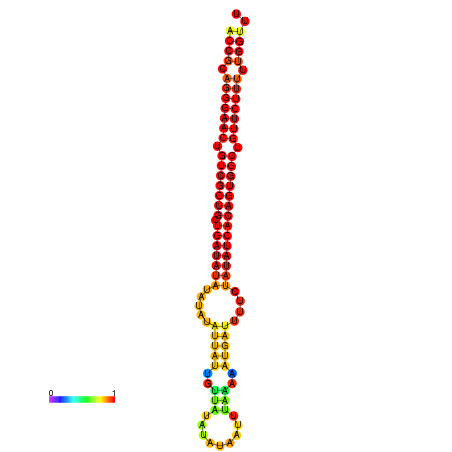 |
 |
| dG=-25.5, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.9, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.7, p-value=0.009901 |
|---|---|---|---|---|
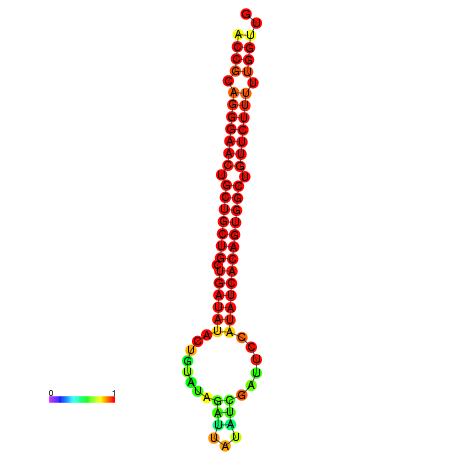 |
 |
 |
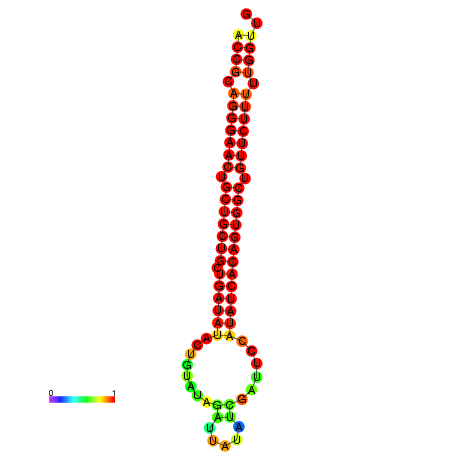 |
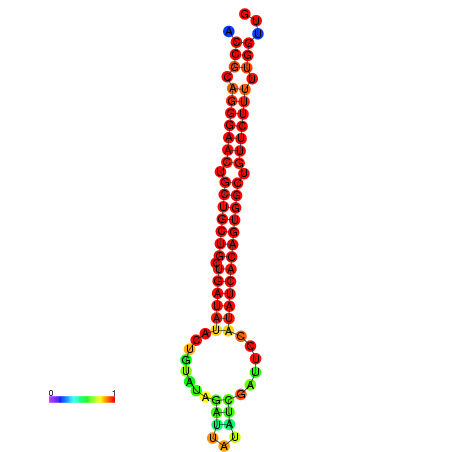 |
| dG=-28.2, p-value=0.009901 | dG=-27.9, p-value=0.009901 | dG=-27.4, p-value=0.009901 |
|---|---|---|
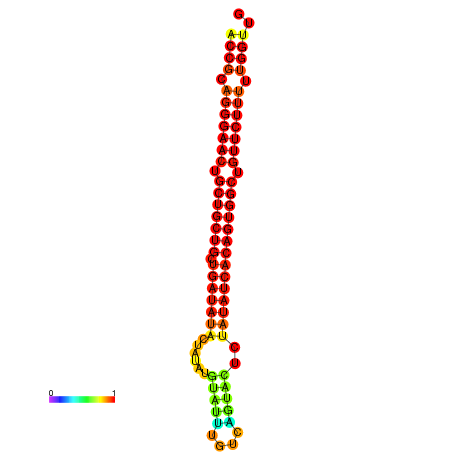 |
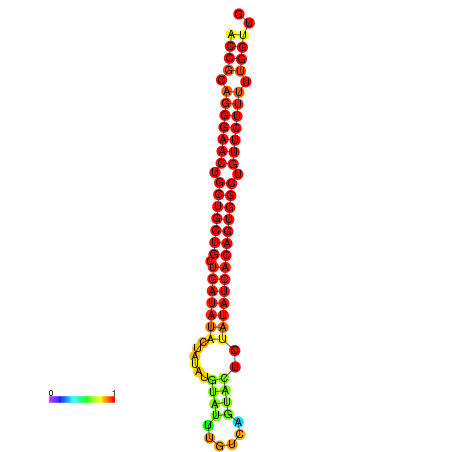 |
 |
| dG=-26.7, p-value=0.009901 | dG=-25.9, p-value=0.009901 |
|---|---|
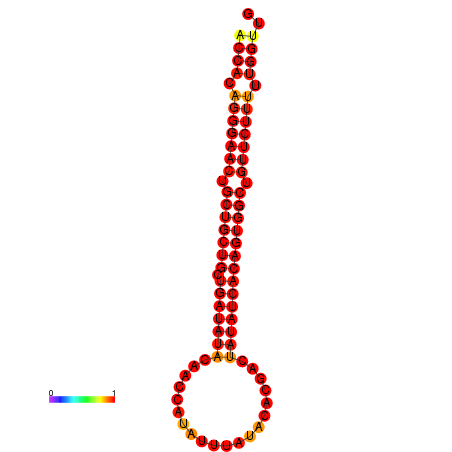 |
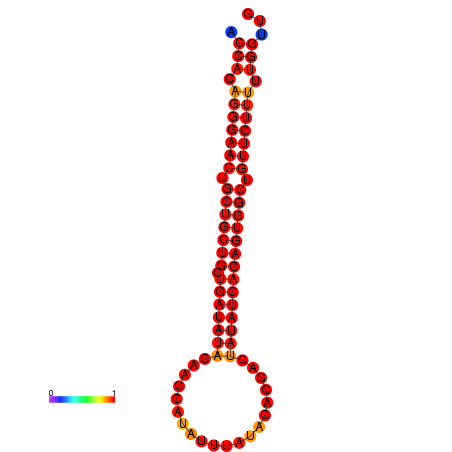 |
Generated: 03/07/2013 at 07:00 PM