

| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548493-15548604 - | AACTTAA-----------------------TCACAGCCTTTAATGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATATACC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CTAACATC----------------ATT |
| droSim1 | chr2R:14253929-14254041 - | AACTTAA-----------------------TCACAGCCTTTAAAGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATATACC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAG-GCATTTTACATC----------------ATT |
| droSec1 | super_1:13063224-13063336 - | AACTTAA-----------------------TCACAGCCTTTAAAGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATATACC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAG-GCATTTTACATC----------------ATT |
| droYak2 | chr2R:13624186-13624297 + | AGTTTAA-----------------------TCACAGCCTTTAACGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATAACAA---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CTAACATC----------------ATT |
| droEre2 | scaffold_4845:9748744-9748855 - | AGCTAAA-----------------------TCGCAGCCTTTAAAGTAGAGGGAATAGTTGCCGTGCTGTAAGTTAATATATC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CTAACATC----------------ATC |
| droAna3 | scaffold_13266:9731810-9731917 + | ---------------------------------CCGCCTTTAAAGTAGAGAGAATAGTGGCTGTGCTGTAAGTTT------C--AAAATCTATATCACAGTGGCTGTTCTTTTTGCACCTAAAGTGC--CAAACAGCCAAACCCGAG------AAT |
| dp4 | chr3:12677072-12677187 - | AC-----------------------------ACCAGCCTTTGAAGTAGAGAGAATAGTTGCTGTGCTATATGTCC----TTCG-ACTCTATATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CAAACAGCCAAG-----GAATCGCGTT |
| droPer1 | super_2:8906485-8906600 + | AC-----------------------------ACCAGCCTTTGAAGTAGAGAGAATAGTTGCTGTGCTATATGTCC----TTCG-ACTCTATATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CAAACAGCCAAG-----GAATCGCGTT |
| droWil1 | scaffold_180701:1651298-1651414 - | T---CAA-----------------------C--CAGCCTTTAAAGTAGAGAGAATAGTTGCTGTGTTTTATGCCC----TTTT-CAATTTCATATCACAGTGGCTGTTCTTTTTGTGCCTAAAGTGC--CCCACAACAACA-----ACAACAACAC |
| droVir3 | scaffold_12875:4112589-4112722 + | GACTTAAAACTATATCGCCCAGTTATCTGTTTATAGCCTTTAAAGTAGAGAGAATAGACGCTGTGCTGTATGTTT----TGCGTATGGTCTATATCACAGTGGCTGTTCTTTTTGTACCTGAAGCGC--CGTACAAC----------------GTT |
| droMoj3 | scaffold_6496:20936294-20936412 - | TATTTATTTCTCTAGTGT-----------TTTTTCGCCTTTAAAGTAGAGAGAATAGATGCTGTG-----AGTATATCTGTT---GCATTTATATCACAGTGGCTGTTCTTTTTGTACCTAAATGGC--CAGACAGC----------------ATT |
| droGri2 | scaffold_5621:3389-3488 + | A----------------------------------GCCTTTAATGTAAAGAGAATAGATGCTGTGCTGTATGTTG----AACGTATTGTCTATATCACAGTGGCTGTTCTTTTTGTACCAAAAGTGC--CGTACAAC----------------AAA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:15548493-15548604 - | AACTTAA-----------------------TCACAGCCTTTAATGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATATACC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CTAACATC----------------ATT |
| droSim1 | chr2R:14253929-14254041 - | AACTTAA-----------------------TCACAGCCTTTAAAGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATATACC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAG-GCATTTTACATC----------------ATT |
| droSec1 | super_1:13063224-13063336 - | AACTTAA-----------------------TCACAGCCTTTAAAGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATATACC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAG-GCATTTTACATC----------------ATT |
| droYak2 | chr2R:13624186-13624297 + | AGTTTAA-----------------------TCACAGCCTTTAACGTAGAGGGAATAGTTGCTGTGCTGTAAGTTAATAACAA---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CTAACATC----------------ATT |
| droEre2 | scaffold_4845:9748744-9748855 - | AGCTAAA-----------------------TCGCAGCCTTTAAAGTAGAGGGAATAGTTGCCGTGCTGTAAGTTAATATATC---ATATCTATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CTAACATC----------------ATC |
| droAna3 | scaffold_13266:9731810-9731917 + | ---------------------------------CCGCCTTTAAAGTAGAGAGAATAGTGGCTGTGCTGTAAGTTT------C--AAAATCTATATCACAGTGGCTGTTCTTTTTGCACCTAAAGTGC--CAAACAGCCAAACCCGAG------AAT |
| dp4 | chr3:12677072-12677187 - | AC-----------------------------ACCAGCCTTTGAAGTAGAGAGAATAGTTGCTGTGCTATATGTCC----TTCG-ACTCTATATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CAAACAGCCAAG-----GAATCGCGTT |
| droPer1 | super_2:8906485-8906600 + | AC-----------------------------ACCAGCCTTTGAAGTAGAGAGAATAGTTGCTGTGCTATATGTCC----TTCG-ACTCTATATATCACAGTGGCTGTTCTTTTTGTACCTAAAGTGC--CAAACAGCCAAG-----GAATCGCGTT |
| droWil1 | scaffold_180701:1651298-1651414 - | T---CAA-----------------------C--CAGCCTTTAAAGTAGAGAGAATAGTTGCTGTGTTTTATGCCC----TTTT-CAATTTCATATCACAGTGGCTGTTCTTTTTGTGCCTAAAGTGC--CCCACAACAACA-----ACAACAACAC |
| droVir3 | scaffold_12875:4112589-4112722 + | GACTTAAAACTATATCGCCCAGTTATCTGTTTATAGCCTTTAAAGTAGAGAGAATAGACGCTGTGCTGTATGTTT----TGCGTATGGTCTATATCACAGTGGCTGTTCTTTTTGTACCTGAAGCGC--CGTACAAC----------------GTT |
| droMoj3 | scaffold_6496:20936294-20936412 - | TATTTATTTCTCTAGTGT-----------TTTTTCGCCTTTAAAGTAGAGAGAATAGATGCTGTG-----AGTATATCTGTT---GCATTTATATCACAGTGGCTGTTCTTTTTGTACCTAAATGGC--CAGACAGC----------------ATT |
| droGri2 | scaffold_5621:3389-3488 + | A----------------------------------GCCTTTAATGTAAAGAGAATAGATGCTGTGCTGTATGTTG----AACGTATTGTCTATATCACAGTGGCTGTTCTTTTTGTACCAAAAGTGC--CGTACAAC----------------AAA |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
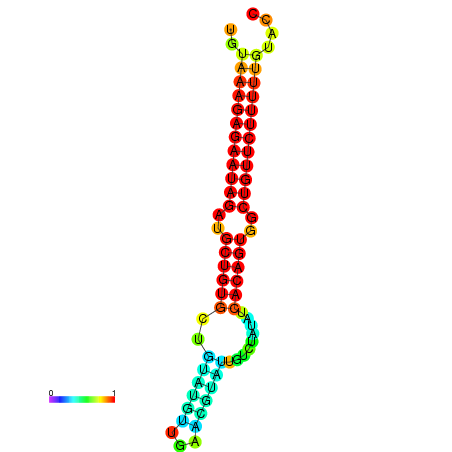
| dG=-24.2, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.4, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.2, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.2, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
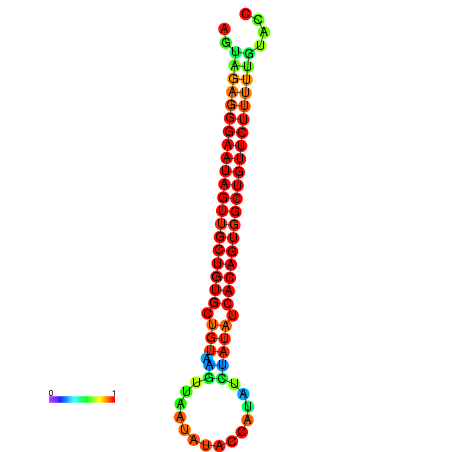 |
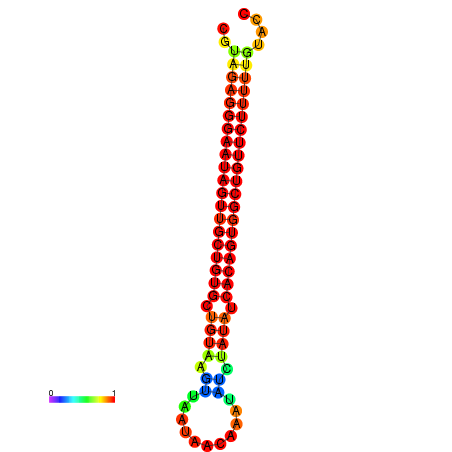
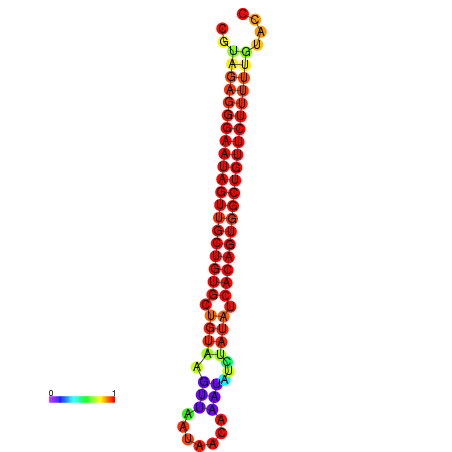
| dG=-24.2, p-value=0.009901 | dG=-24.0, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-23.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
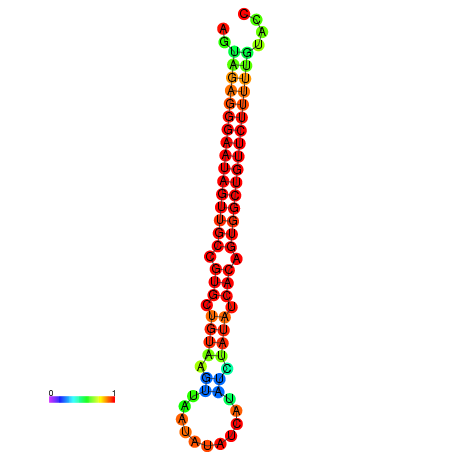
| dG=-19.5, p-value=0.009901 | dG=-19.3, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-18.8, p-value=0.009901 | dG=-18.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
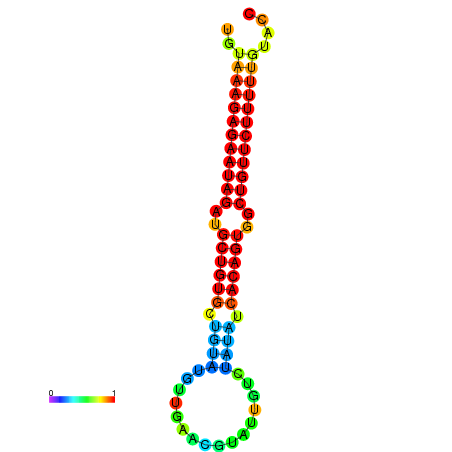
| dG=-30.0, p-value=0.009901 | dG=-29.3, p-value=0.009901 |
|---|---|
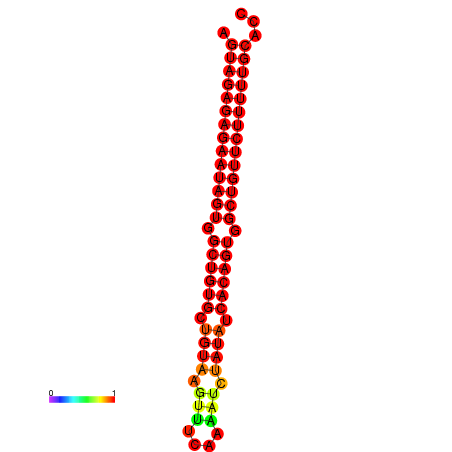 |
 |
| dG=-28.7, p-value=0.009901 | dG=-28.0, p-value=0.009901 |
|---|---|
 |
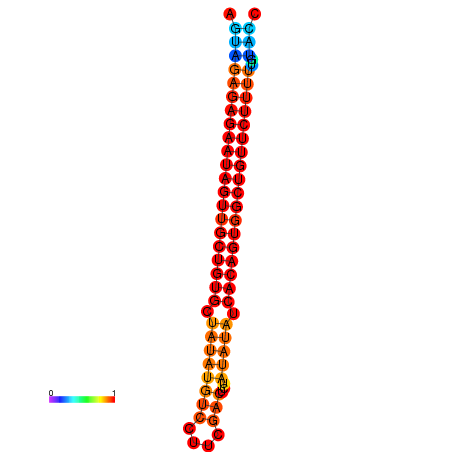 |
| dG=-28.7, p-value=0.009901 | dG=-28.0, p-value=0.009901 |
|---|---|
 |
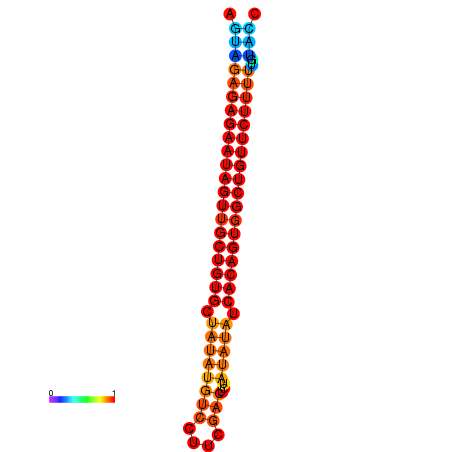 |
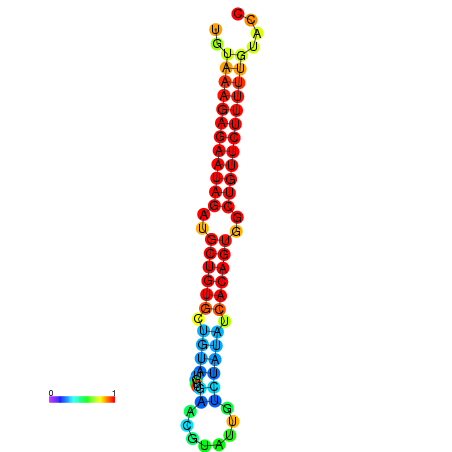
| dG=-23.9, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|
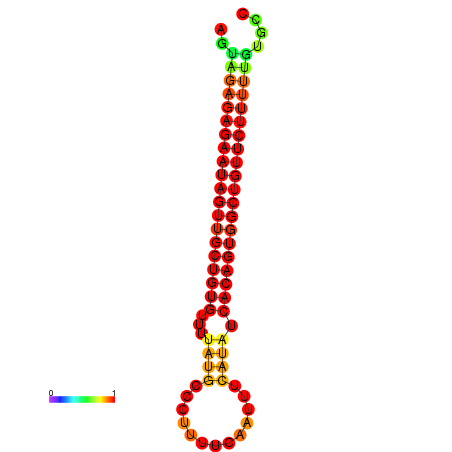 |
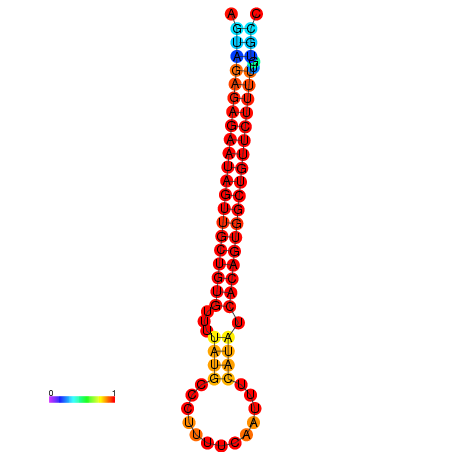 |
| dG=-26.2, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|
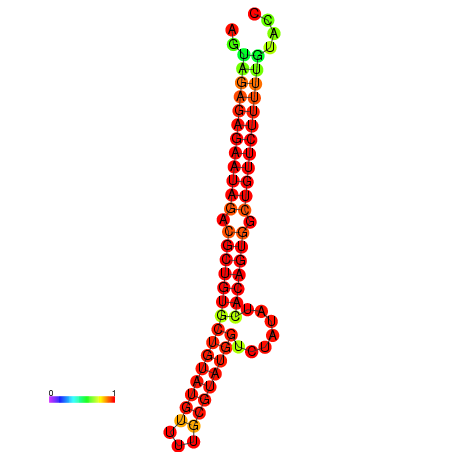 |
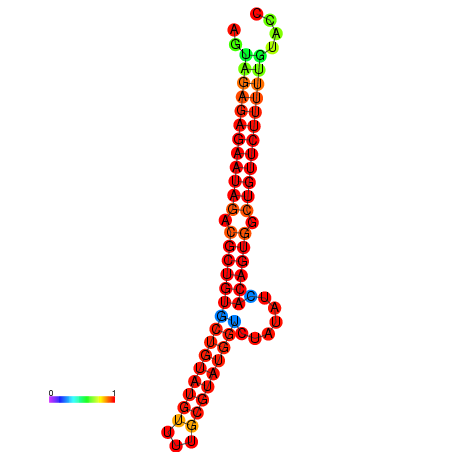 |
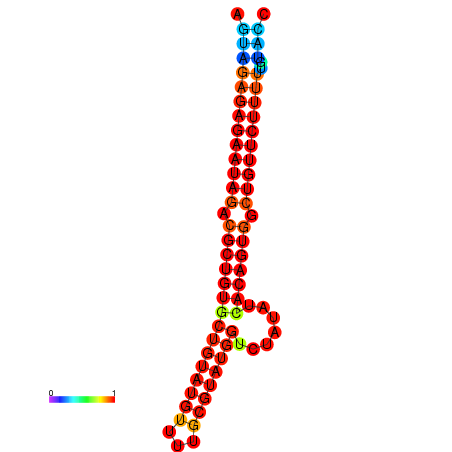 |
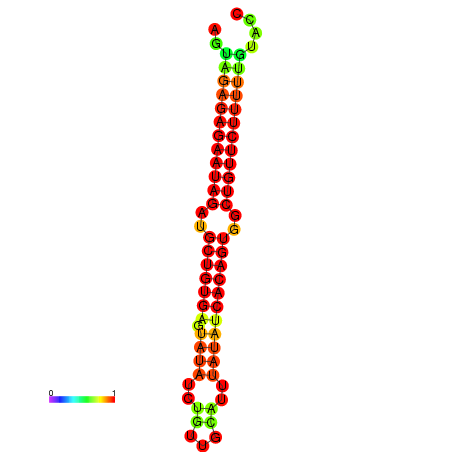
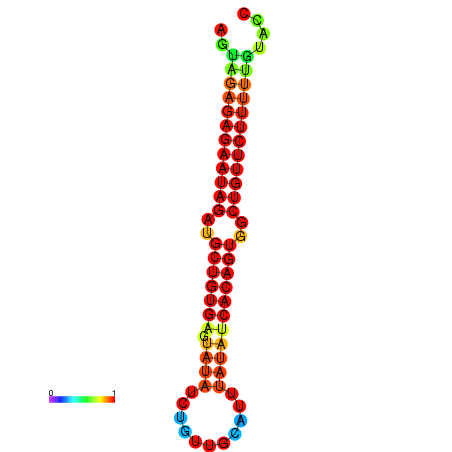
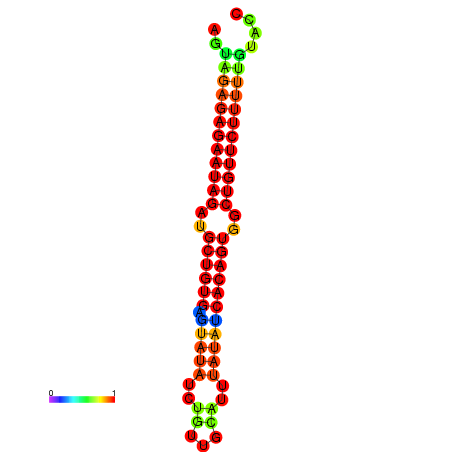
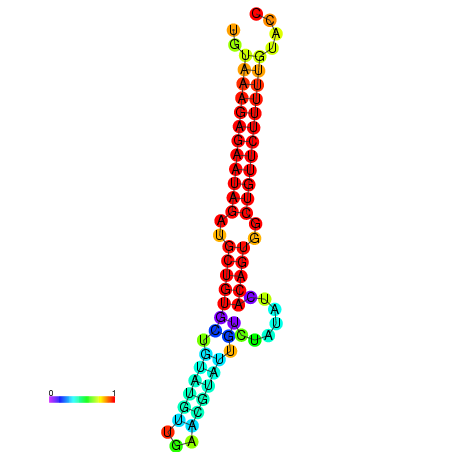
| dG=-23.9, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-23.2, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
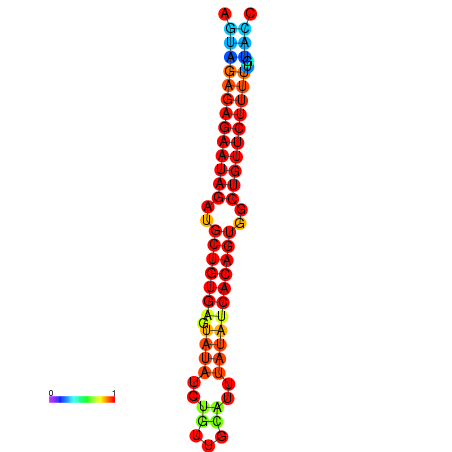 |
| dG=-21.3, p-value=0.009901 | dG=-21.0, p-value=0.009901 | dG=-20.8, p-value=0.009901 | dG=-20.7, p-value=0.009901 | dG=-20.6, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
Generated: 03/07/2013 at 06:59 PM