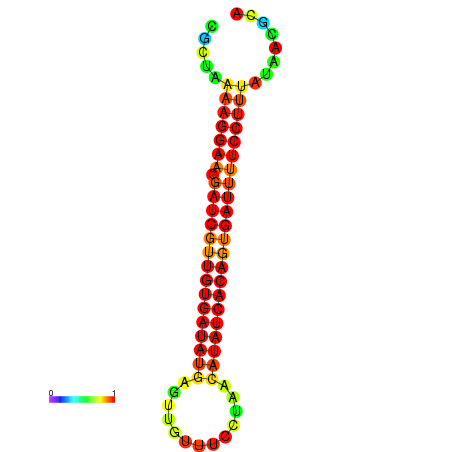
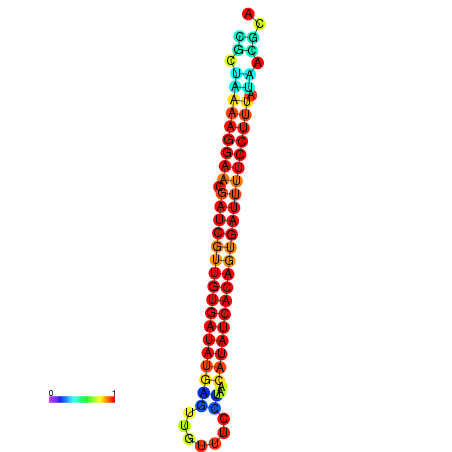
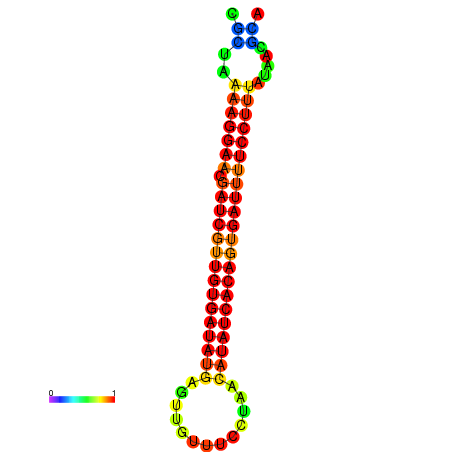
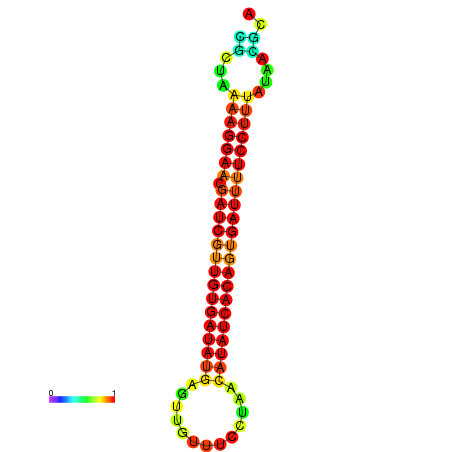
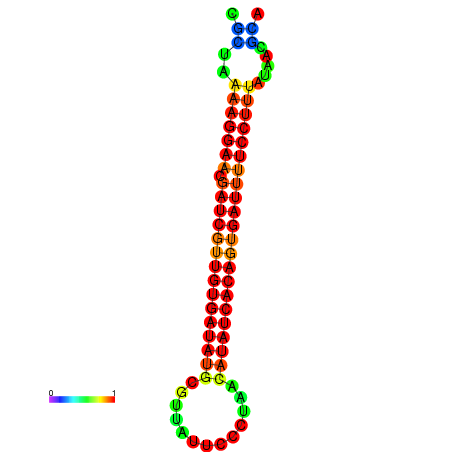
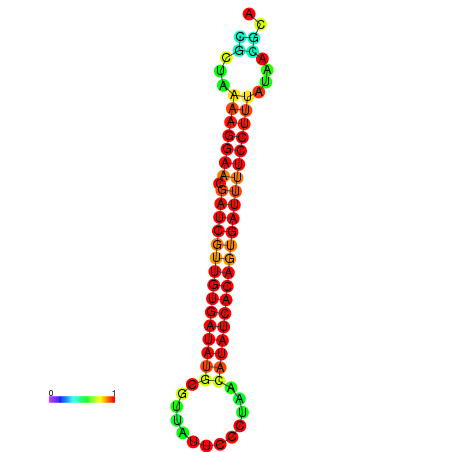
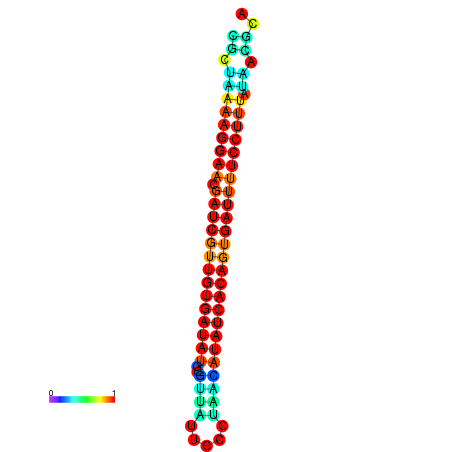
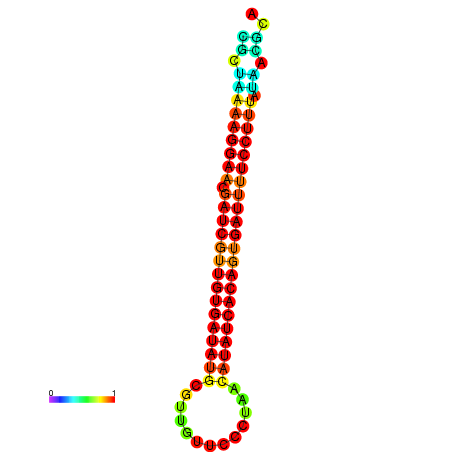
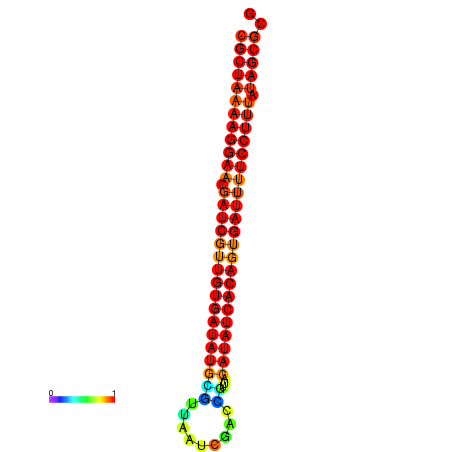
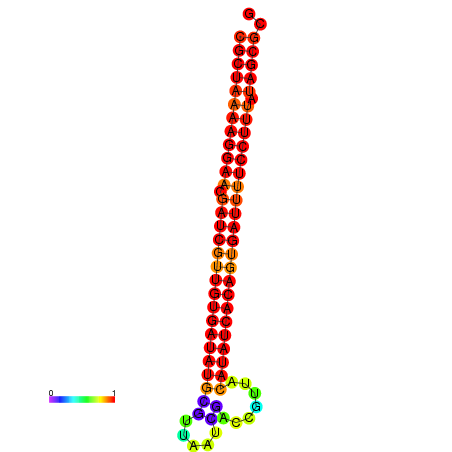
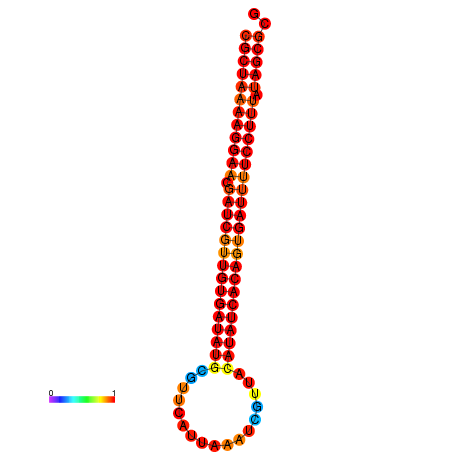| TTCCTAGGGGCCATACGCTAAAAGGAACGATCGTTGTGATATGCGTTAATTCACCGTTACATATCACAGTGATTTTCCTTTATAGCGCATGTCT-GCGCCCAC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ....................AAAGGAACGATCGTTGTGATATG............................................................ | 23 | 1 | 7316.00 | 7316 | 7312 | 4 |
| .............................................................TATCACAGTGATTTTCCTTTAT.................... | 22 | 1 | 3041.00 | 3041 | 3041 | 0 |
| .............................................................TATCACAGTGATTTTCCTTTATA................... | 23 | 1 | 1872.00 | 1872 | 1872 | 0 |
| ....................AAAGGAACGATCGTTGTGATAT............................................................. | 22 | 1 | 1796.00 | 1796 | 1796 | 0 |
| ....................AAAGGAACGATCGTTGTGATA.............................................................. | 21 | 1 | 454.00 | 454 | 454 | 0 |
| ....................AAAGGAACGATCGTTGTGAT............................................................... | 20 | 1 | 266.00 | 266 | 265 | 1 |
| ..............................................................ATCACAGTGATTTTCCTTTAT.................... | 21 | 1 | 196.00 | 196 | 196 | 0 |
| ..............................................................ATCACAGTGATTTTCCTTTATA................... | 22 | 1 | 138.00 | 138 | 138 | 0 |
| .............................................................TATCACAGTGATTTTCCTTT...................... | 20 | 1 | 50.00 | 50 | 50 | 0 |
| .................................................................ACAGTGATTTTCCTTTAT.................... | 18 | 1 | 36.00 | 36 | 36 | 0 |
| ..................TAAAAGGAACGATCGTTGTGAT............................................................... | 22 | 1 | 33.00 | 33 | 33 | 0 |
| .TCCTAGGGGCCATACGCTA................................................................................... | 19 | 1 | 32.00 | 32 | 32 | 0 |
| .............................................................TATCACAGTGATTTTCCTTTA..................... | 21 | 1 | 32.00 | 32 | 32 | 0 |
| .................................................................ACAGTGATTTTCCTTTATA................... | 19 | 1 | 22.00 | 22 | 22 | 0 |
| ....................AAAGGAACGATCGTTGTGATATGC........................................................... | 24 | 1 | 17.00 | 17 | 17 | 0 |
| ...............................................................TCACAGTGATTTTCCTTTAT.................... | 20 | 1 | 15.00 | 15 | 15 | 0 |
| TTCCTAGGGGCCATACGCTA................................................................................... | 20 | 1 | 14.00 | 14 | 14 | 0 |
| ..................TAAAAGGAACGATCGTTGTGATA.............................................................. | 23 | 1 | 13.00 | 13 | 13 | 0 |
| ...............................................................TCACAGTGATTTTCCTTTATA................... | 21 | 1 | 13.00 | 13 | 13 | 0 |
| ....................AAAGGAACGATCGTTGTGA................................................................ | 19 | 1 | 13.00 | 13 | 13 | 0 |
| .............................................................TATCACAGTGATTTTCCT........................ | 18 | 1 | 5.00 | 5 | 5 | 0 |
| .........................................TGCGTTAATTCACCGTTACATA........................................ | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ................................................................CACAGTGATTTTCCTTTAT.................... | 19 | 1 | 4.00 | 4 | 4 | 0 |
| ....................AAAGGAACGATCGTTGTG................................................................. | 18 | 1 | 4.00 | 4 | 4 | 0 |
| .....................AAGGAACGATCGTTGTGATATG............................................................ | 22 | 1 | 3.00 | 3 | 3 | 0 |
| ...............................................................TCACAGTGATTTTCCTTTATAGC................. | 23 | 1 | 3.00 | 3 | 3 | 0 |
| .............................................................TATCACAGTGATTTTCCTT....................... | 19 | 1 | 2.00 | 2 | 2 | 0 |