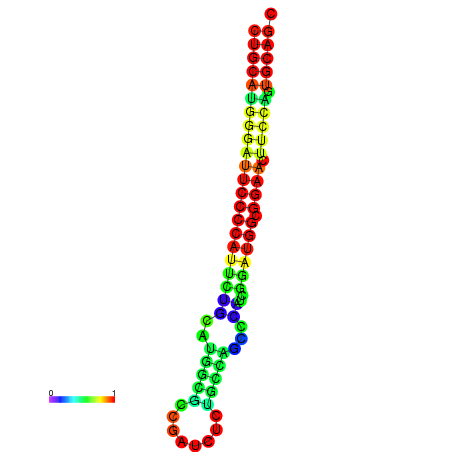
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:9726743-9726895 - | AGACTGCCAACTGGGTTC------TGCCGCTT---TTGCTGTGGCTTCTCTGCATGGGATTCCCCATTCTGCATGGCGCCGATCTCTGCCAGCCCATCGGATGGCGGAACTTCCAGTGCAGCGAGGTCGCTTCTCTGCAGGATCTGGTGGACTTGGGCG---------CCG |
| droSim1 | chr3L:9107366-9107518 - | CAACTGCCAACTGGGTTC------TGCCGCTT---TTGCTGTGGCTTCTCTGCGTGGGATTCCCCATCCTGCATGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGGTGGATTTGGGCG---------CCG |
| droSec1 | super_0:1960228-1960380 - | CAACTGCCAACTGGGTTC------TGTCGCTT---TTGCTGTGCCTTCTCTGCGTGGGATTCCCCATCCTGCATGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGCAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGGTGGACCTGGGCG---------CCG |
| droYak2 | chr3L:9698225-9698377 - | CAACTGCCAATTGGGTTC------TGCCGCTT---TTTCTGTGGCTTCTCTGCGCGGGATTCCCCAGCCTGCATGGTGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGAATTGGGCG---------CCG |
| droEre2 | scaffold_4784:9713780-9713932 - | CAACTGCCAATTGGGTTC------TGCCGCTC---TTTCTGTGCCTTCTCTGCGCGGGATTCCCCAGCCTGCATGGAGCTGATCTCTGCCAGCCCGTCGGGTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGACCTGGGCG---------CCG |
| droAna3 | scaffold_13337:3652327-3652482 + | TGCCAGTCAAATGGACTC------TGTTGCCTTGCTTTTTGTGGCTCTTCTCGAAGGGGCTTCCCTCCACAAGAGCTCAAGATCTTTGCCAGCCCCGAGGTTGGCGCAACTTCGAGTGCCTTGAAGTGGCTTCTCTGCAGGATCTAGTGGATTTGGGGG---------CCG |
| dp4 | chrXR_group6:9932877-9933023 + | TGAATGCCAATTGGGTTT------TGTCC------GCTCTGTGGCTCTGCTGCAGGGGCTTCCCCC---AGGTCGGGGCTCTTCCCTGCCGGACTCTGGCCTGGCGCAGCTTCGAGTGTCGTGAATTAGCCAGTCTGCAGGATCTGTCTGATGTGGGGG---------CCG |
| droPer1 | super_61:212952-213098 + | TGAATGCCAATTGGGTTT------TGTCC------GCTCTGTGGCTCTGCTGCAGGGGCTTCCCCC---AGGTCGGGGCTCTTCCCTGCCGGACTCTGGCCTGGCGCAGCTTCGAGTGTCGTGAATTAGCCAGTCTGCAGGATCTGTCTGATGTGGGGG---------CCG |
| droWil1 | scaffold_180949:1195301-1195459 + | CGAATTTCCAGTGGTTTT------GGCCGTTG---GCTTTATGGCTTGTATGTCGGTGCTTCCCGC---GGTGCACAGCTCAAGTCTGTAATCCCCGCGGCTGGCGTAACTACGAATGCAGCGAATTGGCTAGTCTAGAGGATTTAATAAATTCTGAAGGCTTGTCCACTG |
| droVir3 | scaffold_13049:8736379-8736540 + | CGAATGCCAAACAGTTCTGGACTTGG---TCA---GTTCTCTGGCTGCTCTGCTGGGCTGTGCCCC---GTGCGTTGGCGCAGTTGTGCGAGGCGCAGGGCTGGCGCAGCTTTGAGTGCCGCGAGCTGGGCAGCCTGCAGGAGCTGATCAATGAGAAGGCCTTGCCCACTG |
| droMoj3 | scaffold_6680:6608267-6608428 - | TGAATGCGCAACCTTTGCGCCTAGGT---CCA---GTCCTCTGGCTGCTGTGCTGCGGCCTTTCCC---AGGTGCTGAGTCAGATATGCGAGGCTCAAGGCTGGCGCAGCTTCGAGTGTCGTGAGCTGAGCAGCCTGAAGGAGCTGAAGAACGTTAGGAACTTTCCCAAAG |
| droGri2 | scaffold_15110:4482444-4482593 + | TGTTCTACACTTGG---------------CCA---ATTCTTTGCCTGCTCTGCTTTGGGTTTTGC--CCTGGAT-TGGCTGTGGTCTGTGAGGCTCAAGGCTGGCGTAGTTTTGAGTGCCGCGAATTGAGCAGCCTGCAGGAGCTGATCGATGTGAAGGCATTTCCCACCG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:9726743-9726895 - | AGACTGCCAACTGGGTTCTGCCGCTTTTGCTGTGGCTTCTCTGCATGGGATTCCCCATTCTGCATGGCGCCGATCTCTGCCAGCCCATCGGATGGCGGAACTTCCAGTGCAGCGAGGTCGCTTCTCTGCAGGATCTGGTGGACTTGGGCGCCG |
| droSim1 | chr3L:9107366-9107518 - | CAACTGCCAACTGGGTTCTGCCGCTTTTGCTGTGGCTTCTCTGCGTGGGATTCCCCATCCTGCATGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGGTGGATTTGGGCGCCG |
| droSec1 | super_0:1960228-1960380 - | CAACTGCCAACTGGGTTCTGTCGCTTTTGCTGTGCCTTCTCTGCGTGGGATTCCCCATCCTGCATGGCGCCGATCTCTGCCAGCCCATCGGCTGGCGCAACTTCCAGTGCAACGAAGTCGCCTCGCTGCAGGATCTGGTGGACCTGGGCGCCG |
| droYak2 | chr3L:9698225-9698377 - | CAACTGCCAATTGGGTTCTGCCGCTTTTTCTGTGGCTTCTCTGCGCGGGATTCCCCAGCCTGCATGGTGCCGATCTCTGCCAGCCCATCGGCTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGAATTGGGCGCCG |
| droEre2 | scaffold_4784:9713780-9713932 - | CAACTGCCAATTGGGTTCTGCCGCTCTTTCTGTGCCTTCTCTGCGCGGGATTCCCCAGCCTGCATGGAGCTGATCTCTGCCAGCCCGTCGGGTGGCGGAACTTCCAGTGCAACGAGGTCGCTTCTCTGCAGGATCTGGTCGACCTGGGCGCCG |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
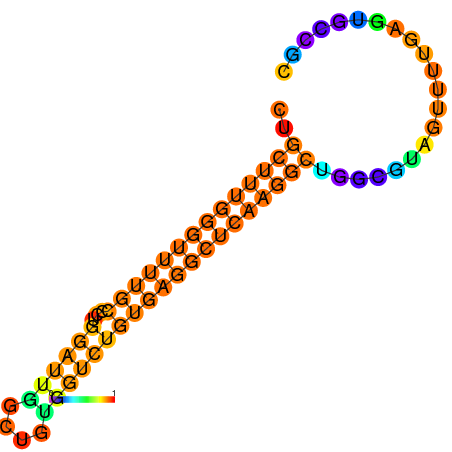
| dG=-30.9, p-value=0.009901 | dG=-30.5, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.2, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
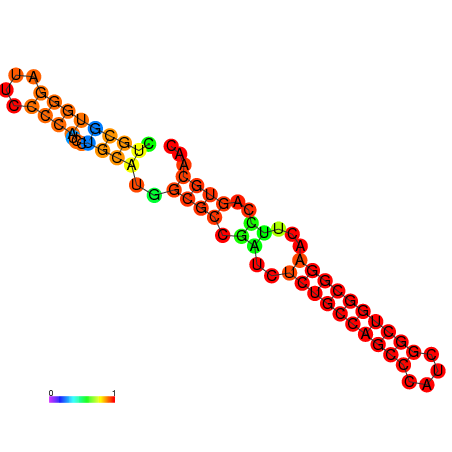
| dG=-32.1, p-value=0.009901 | dG=-31.6, p-value=0.009901 | dG=-31.6, p-value=0.009901 | dG=-31.1, p-value=0.009901 | dG=-31.1, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|---|
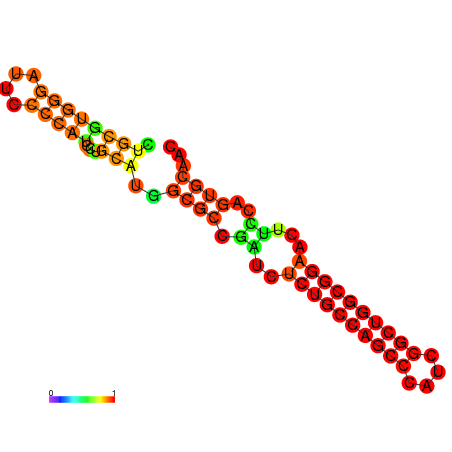 |
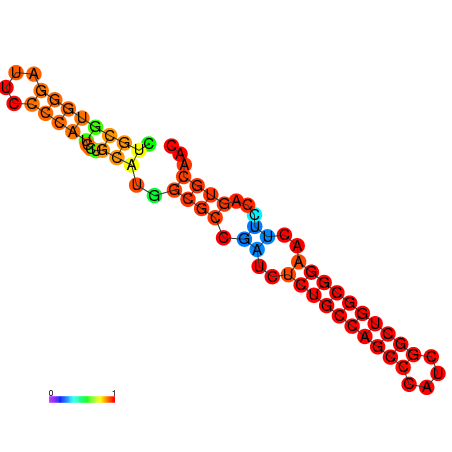 |
 |
 |
 |
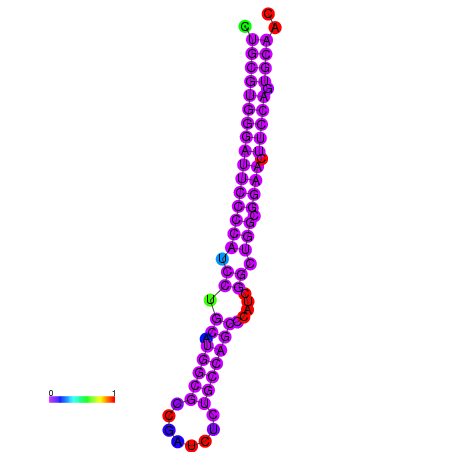 |
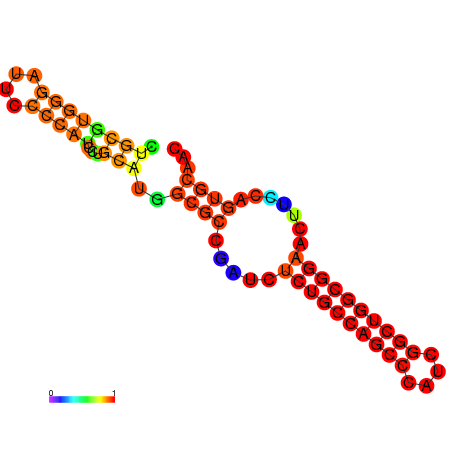
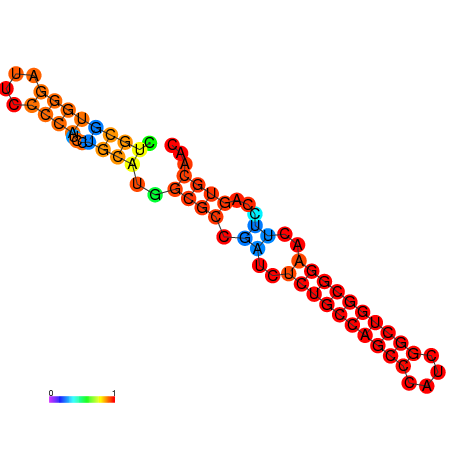
| dG=-27.5, p-value=0.009901 | dG=-27.0, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-26.5, p-value=0.009901 | dG=-19.7, p-value=0.009901 |
|---|---|---|---|---|
 |
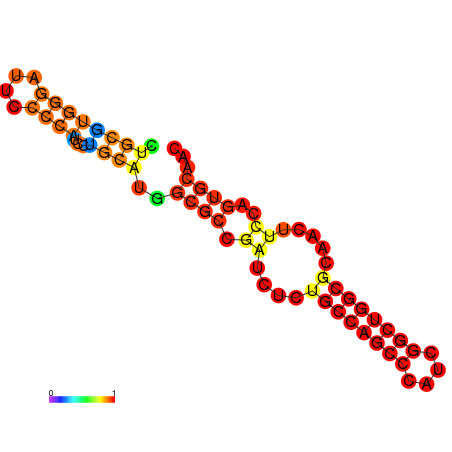 |
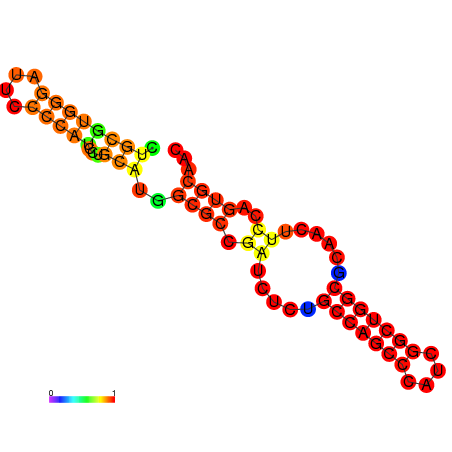 |
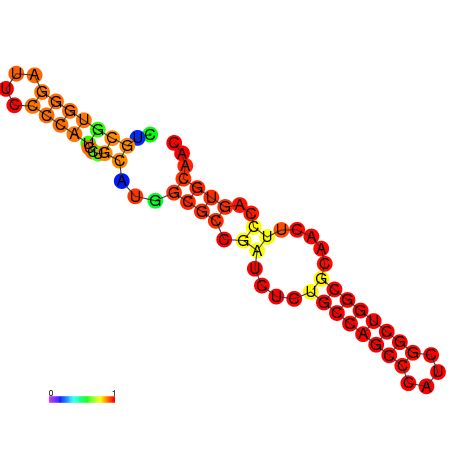 |
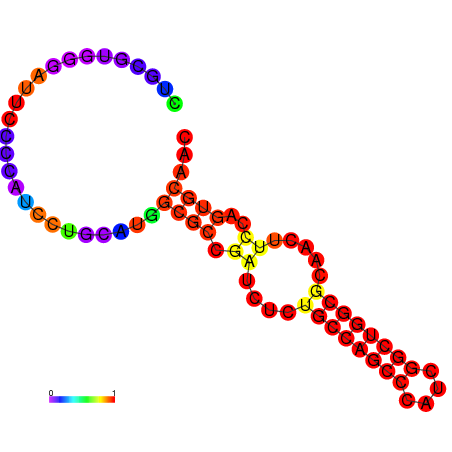 |
| dG=-32.6, p-value=0.009901 | dG=-32.0, p-value=0.009901 |
|---|---|
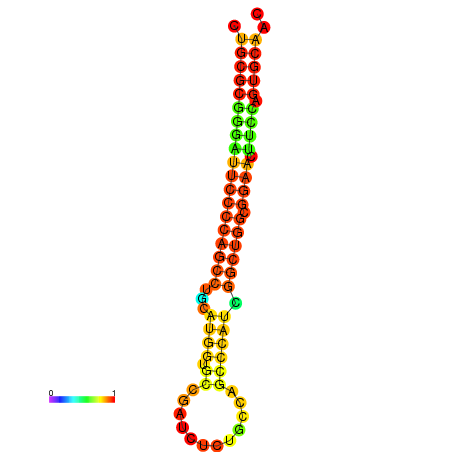 |
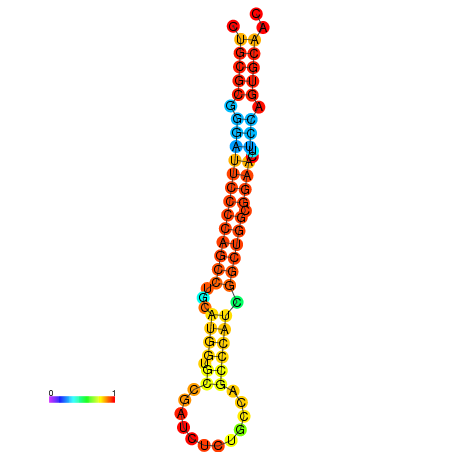 |
| dG=-36.1, p-value=0.009901 | dG=-35.7, p-value=0.009901 | dG=-35.5, p-value=0.009901 | dG=-35.1, p-value=0.009901 |
|---|---|---|---|
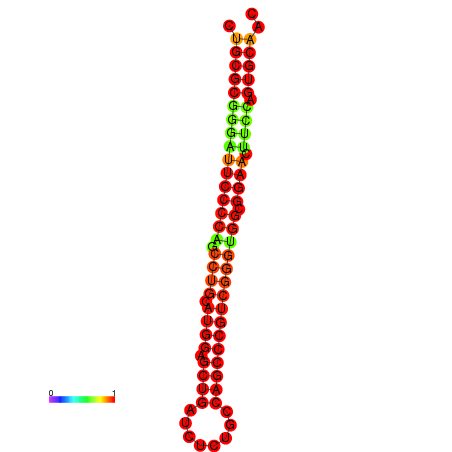 |
 |
 |
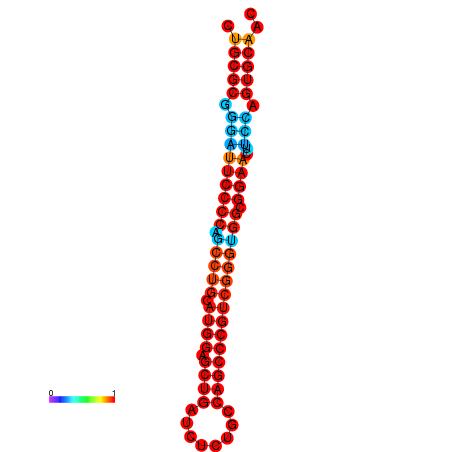 |
| dG=-26.1, p-value=0.009901 | dG=-25.7, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.3, p-value=0.009901 | dG=-23.1, p-value=0.009901 |
|---|---|---|---|---|
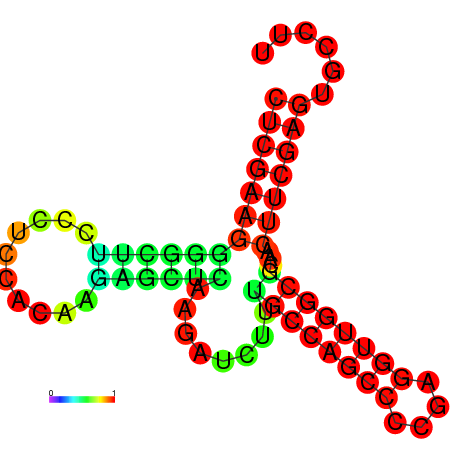 |
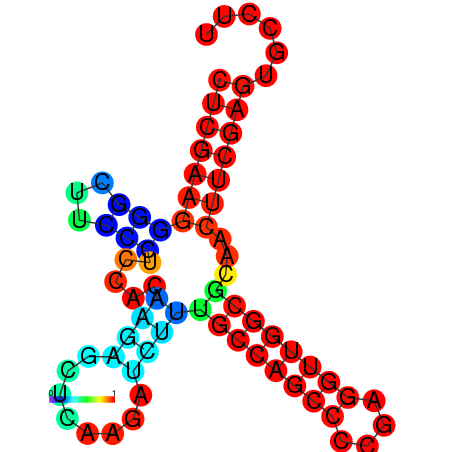
 |
 |
 |
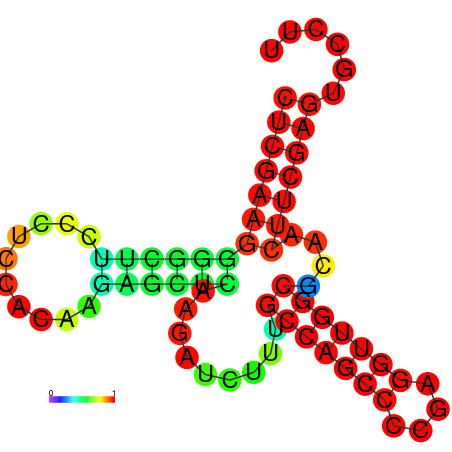
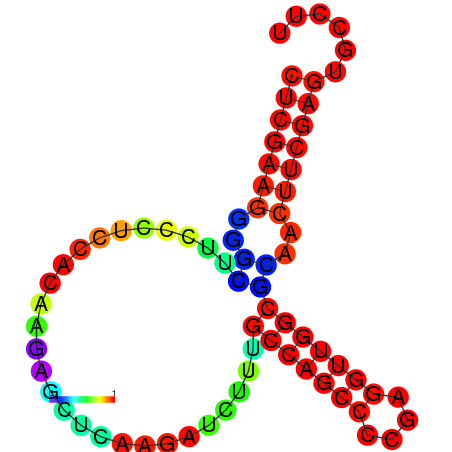 |
| dG=-27.5, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.0, p-value=0.009901 |
|---|---|---|---|---|
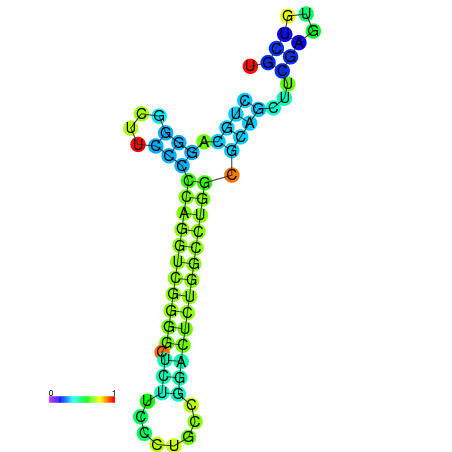 |
 |
 |
 |
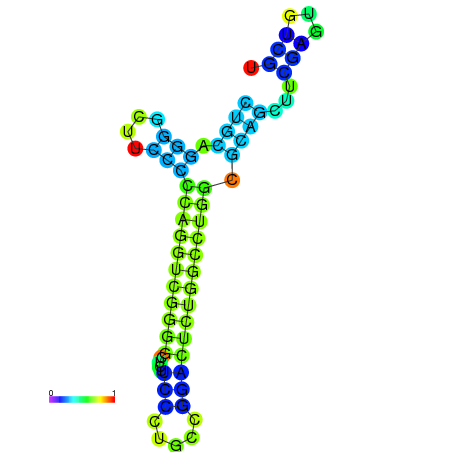 |
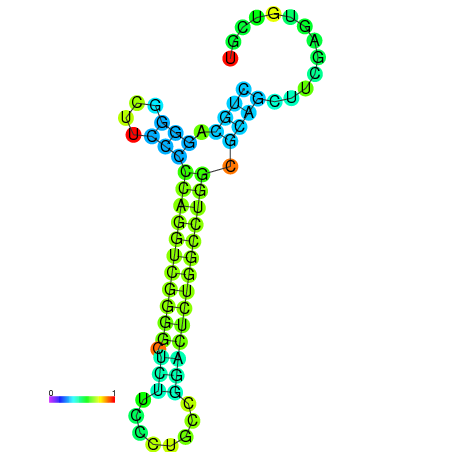
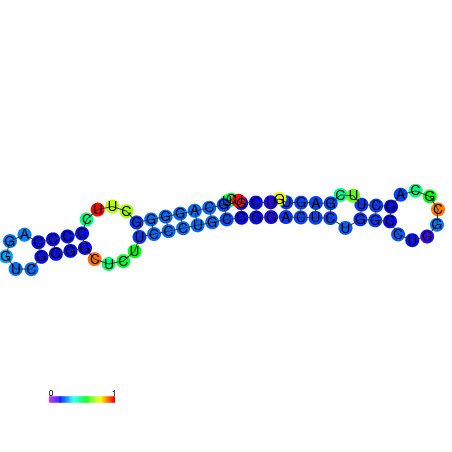
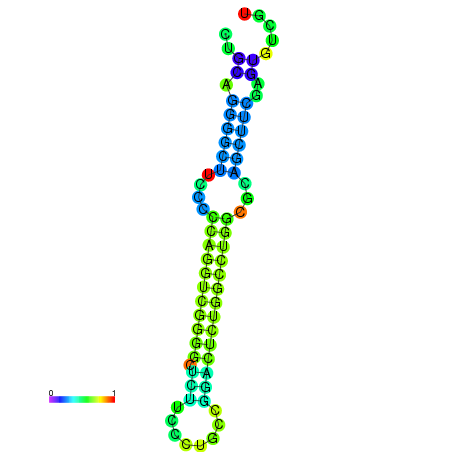
| dG=-27.5, p-value=0.009901 | dG=-27.4, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-27.0, p-value=0.009901 |
|---|---|---|---|---|
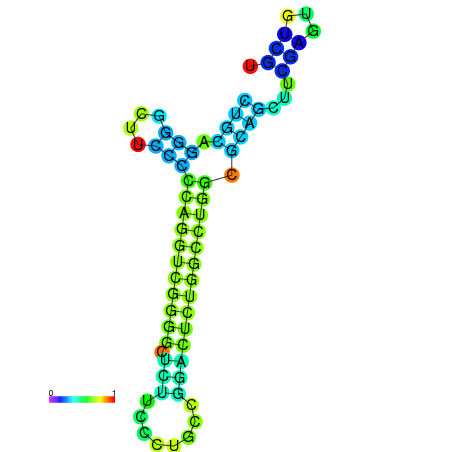 |
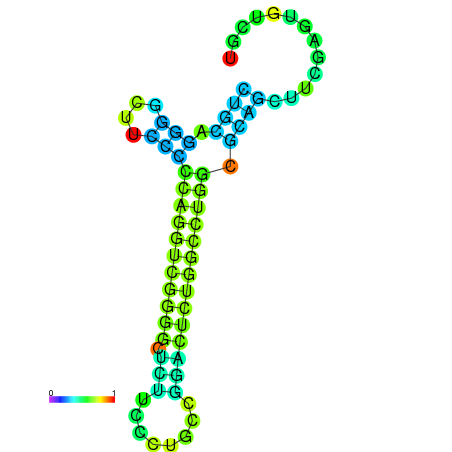 |
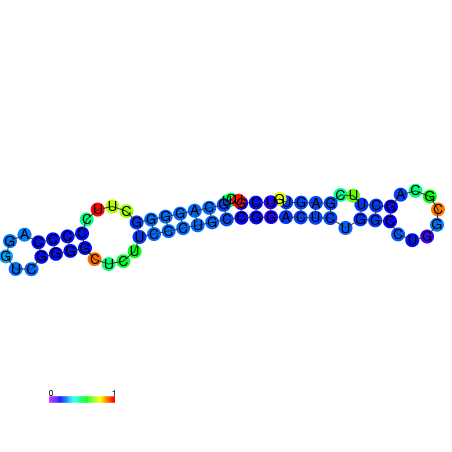 |
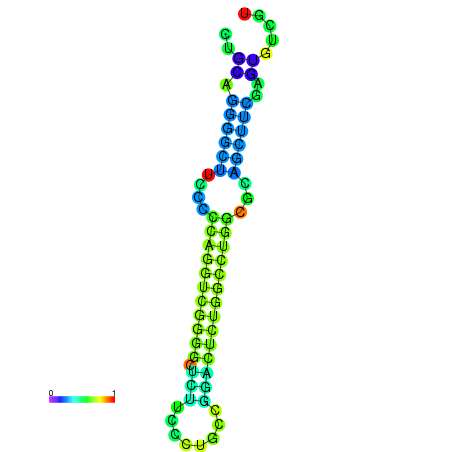 |
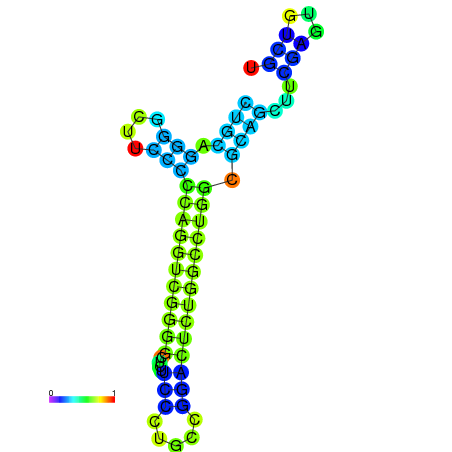 |
| dG=-17.7, p-value=0.009901 | dG=-17.0, p-value=0.009901 | dG=-16.8, p-value=0.009901 | dG=-16.7, p-value=0.009901 |
|---|---|---|---|
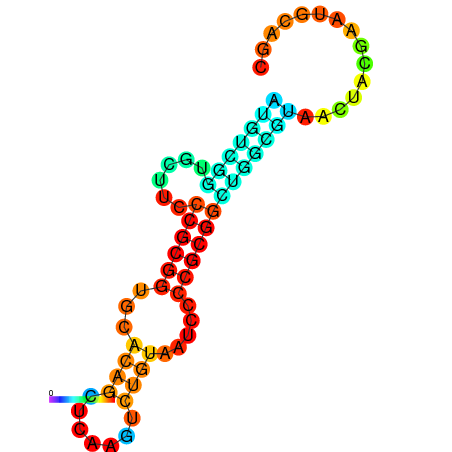 |
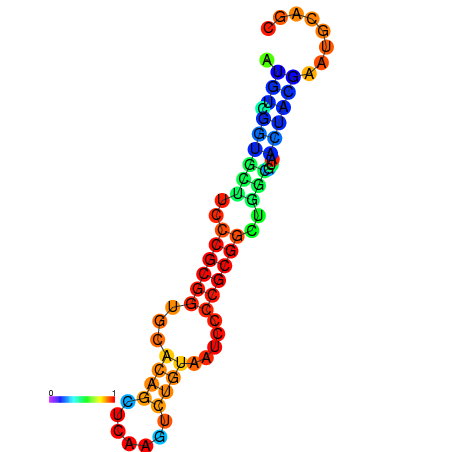 |
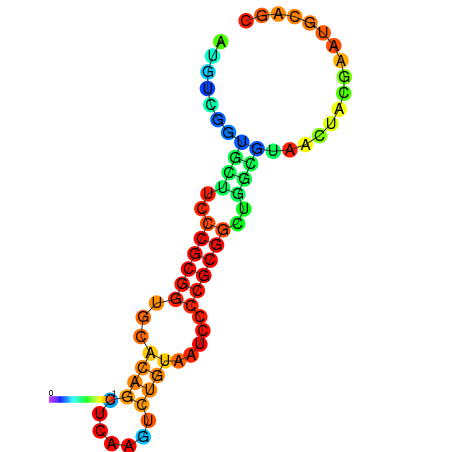 |
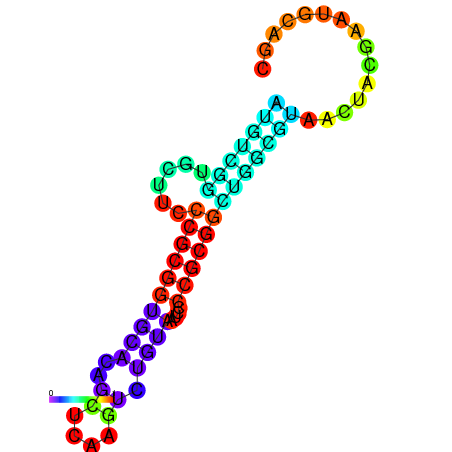 |
| dG=-36.0, p-value=0.009901 | dG=-35.8, p-value=0.009901 | dG=-35.7, p-value=0.009901 | dG=-35.6, p-value=0.009901 | dG=-35.5, p-value=0.009901 |
|---|---|---|---|---|
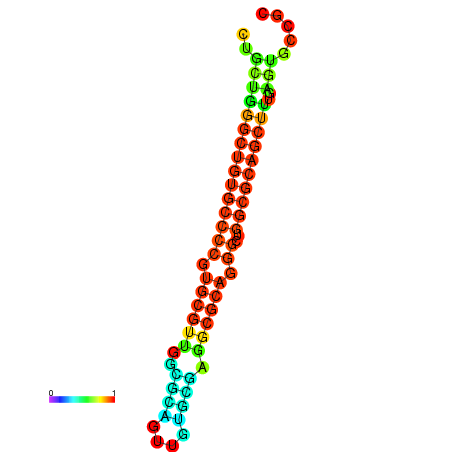 |
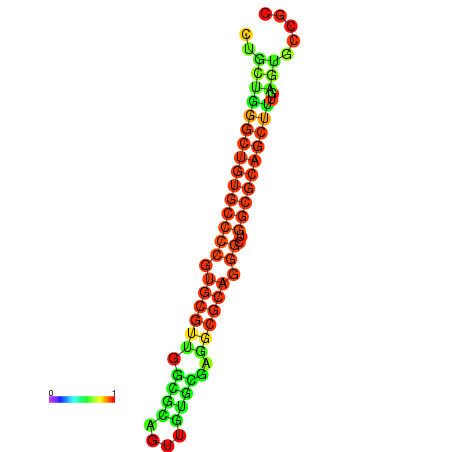 |
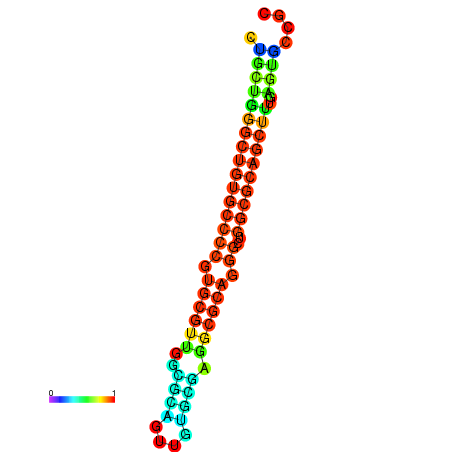 |
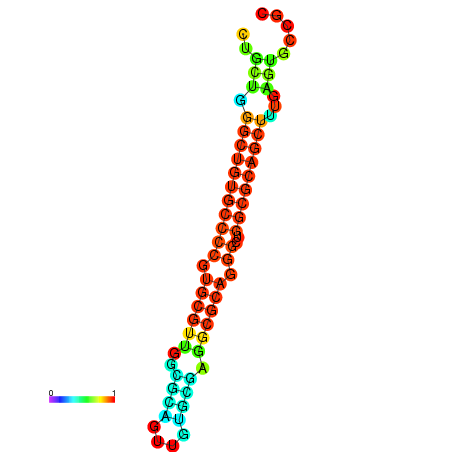 |
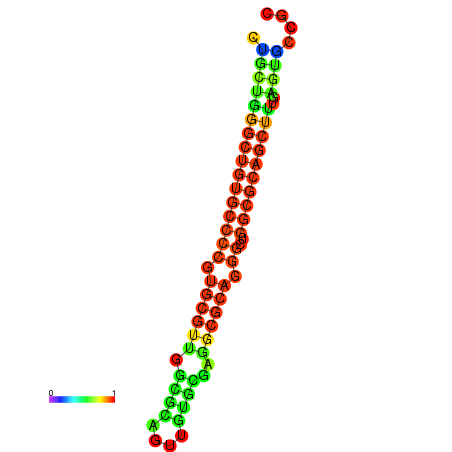 |
| dG=-25.5, p-value=0.009901 | dG=-25.2, p-value=0.009901 | dG=-25.1, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 | dG=-20.8, p-value=0.009901 |
|---|---|---|---|---|---|
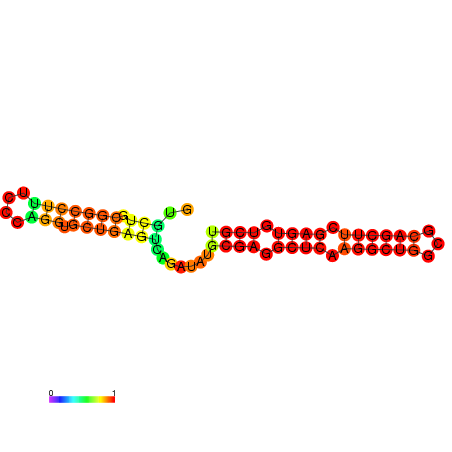 |
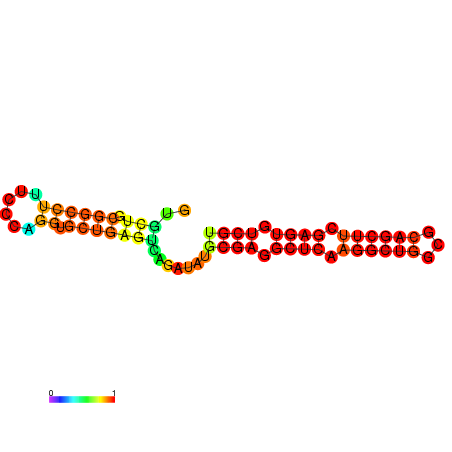 |
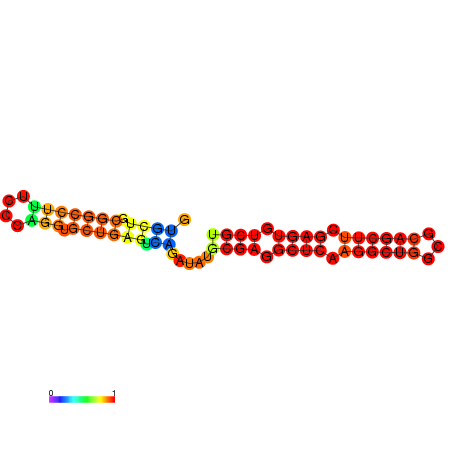 |
 |
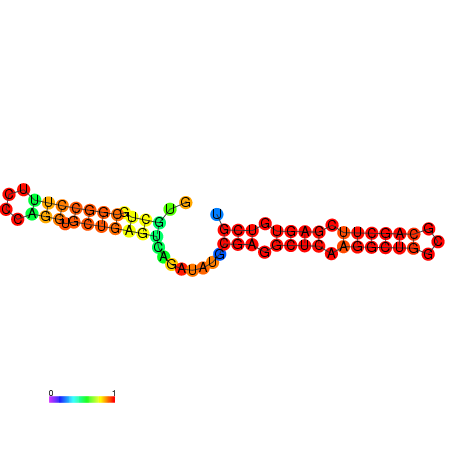 |
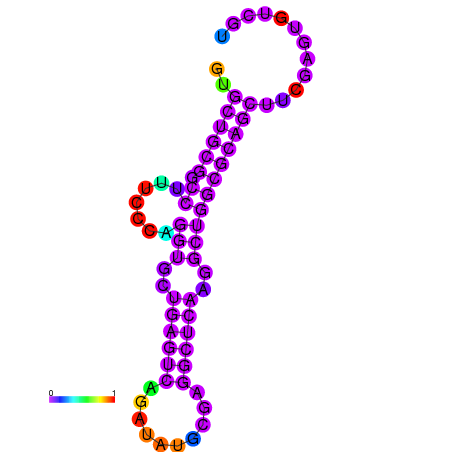 |
| dG=-21.7, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.3, p-value=0.009901 | dG=-18.4, p-value=0.009901 |
|---|---|---|---|---|---|
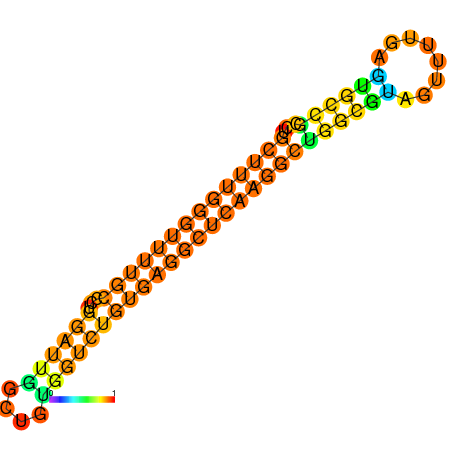 |
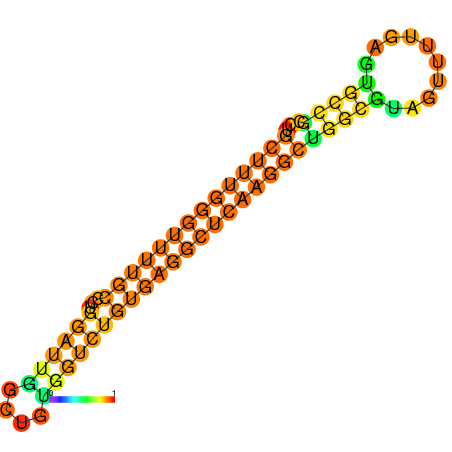 |
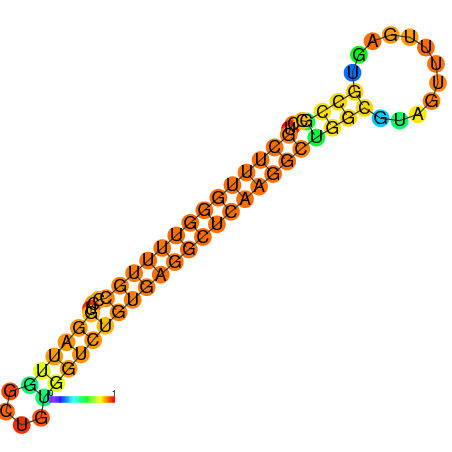 |
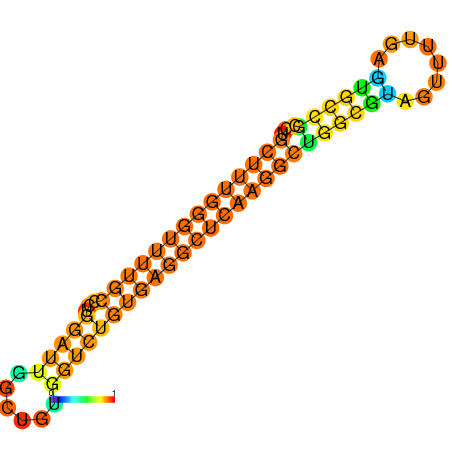 |
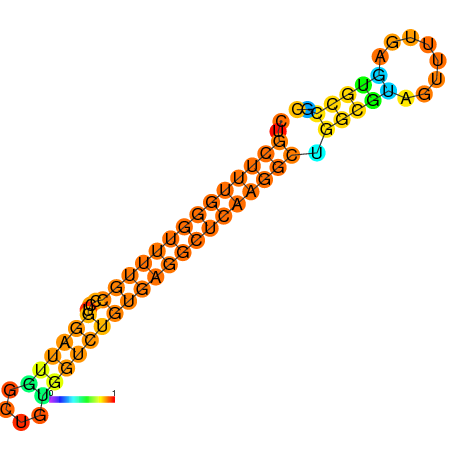 |
 |
Generated: 03/07/2013 at 06:55 PM