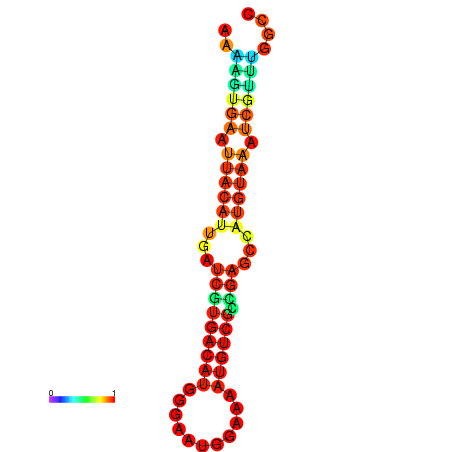
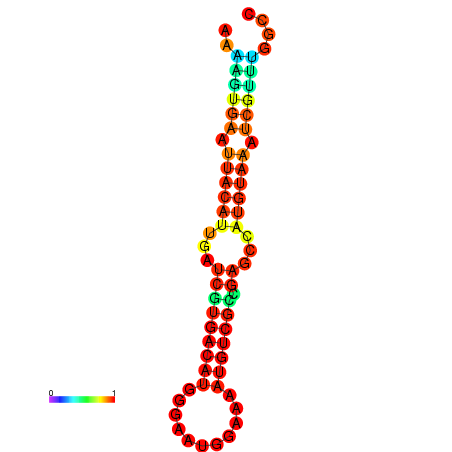
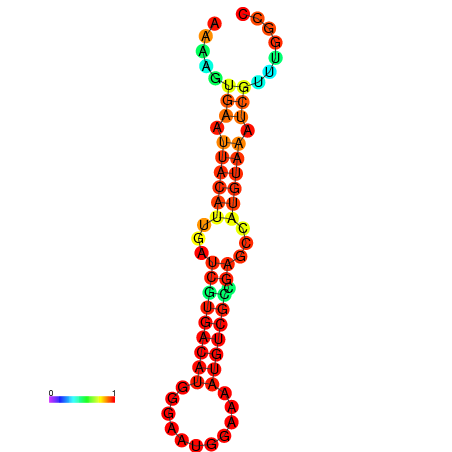
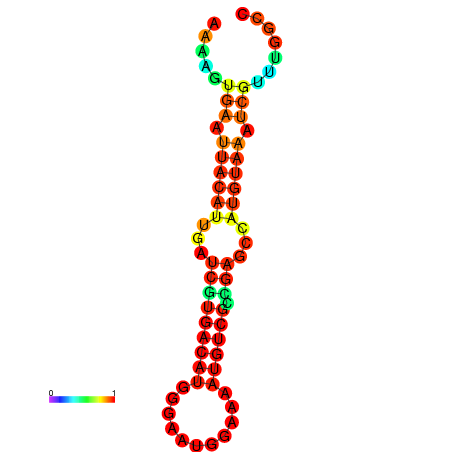
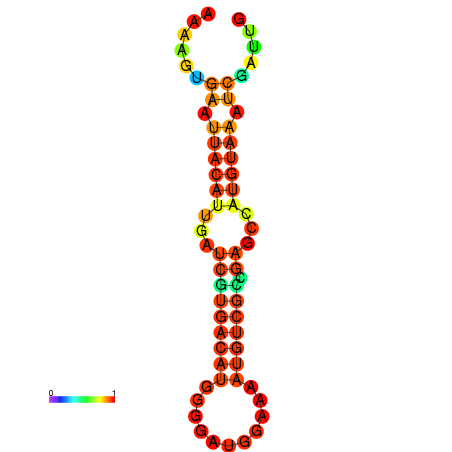
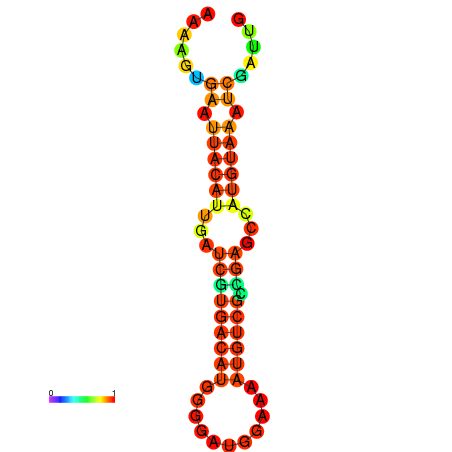
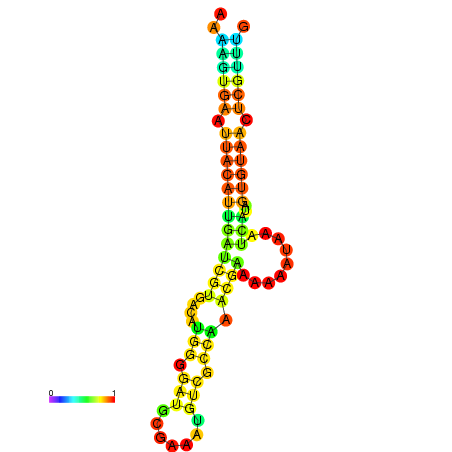
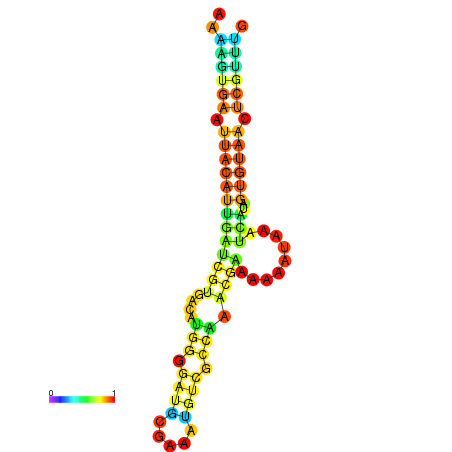
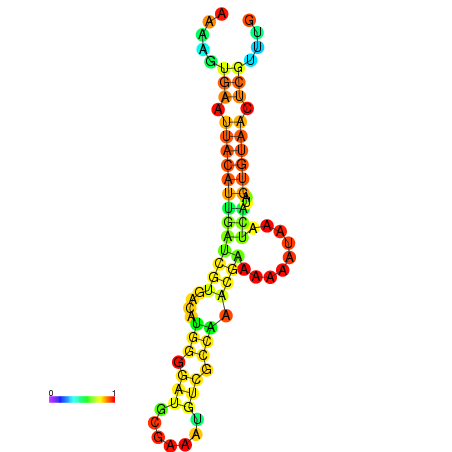
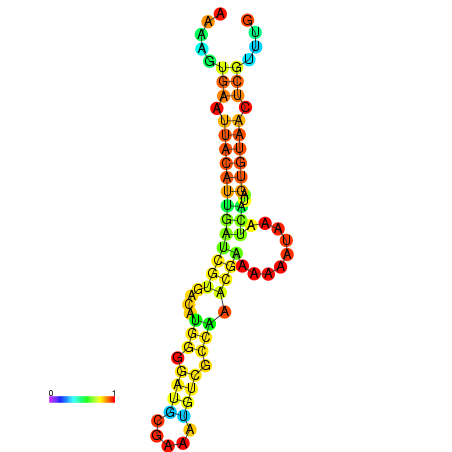
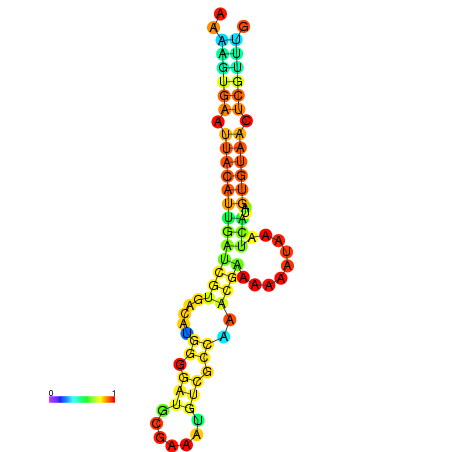
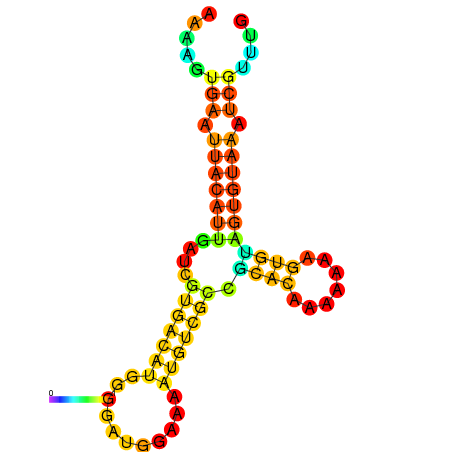
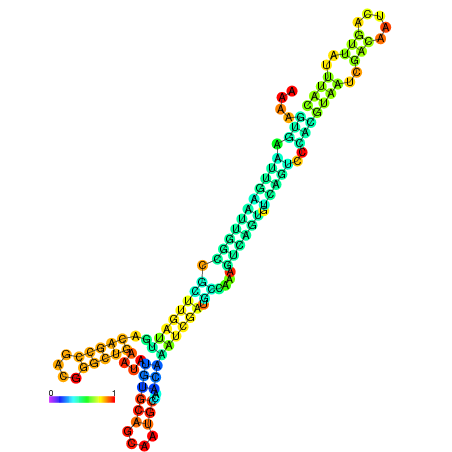
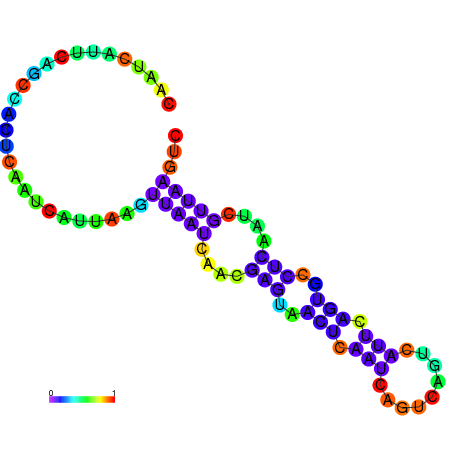
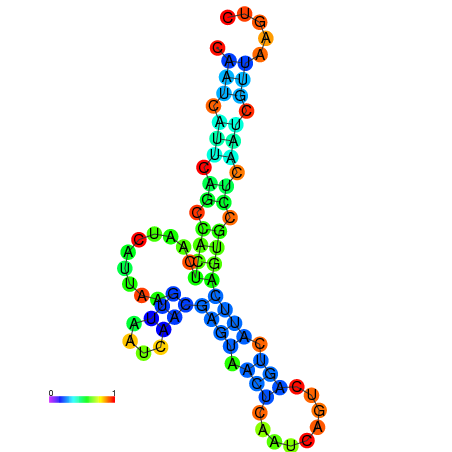
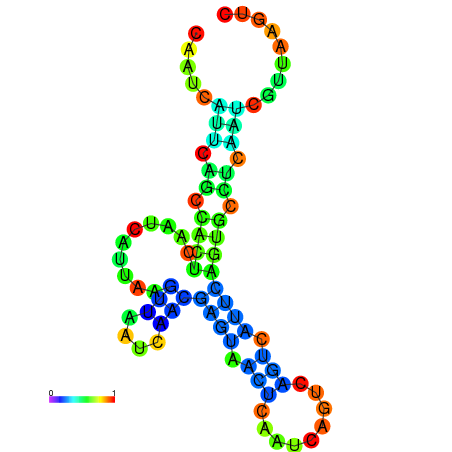
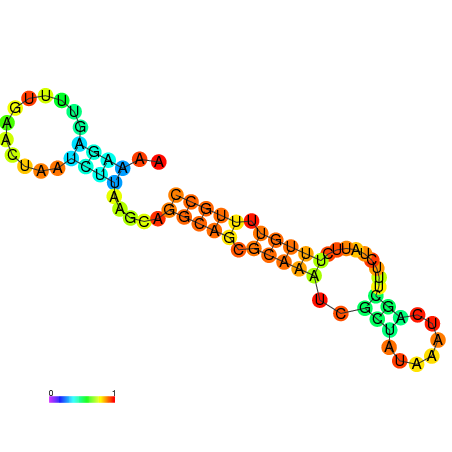
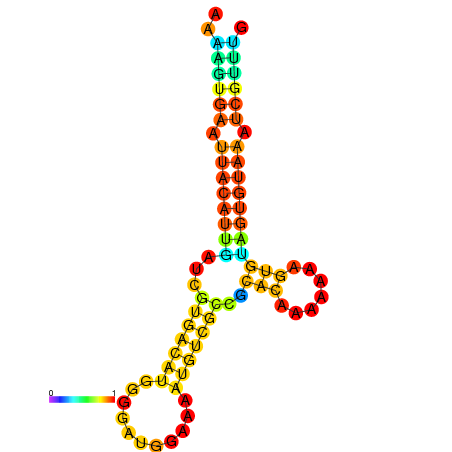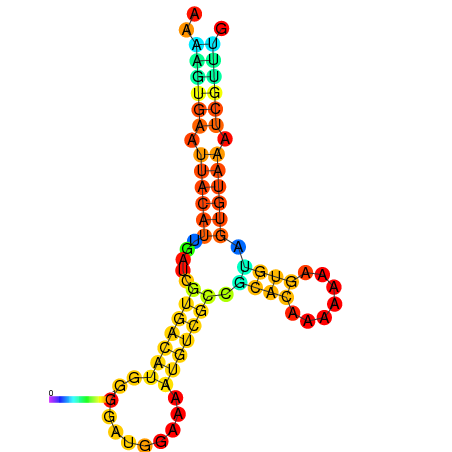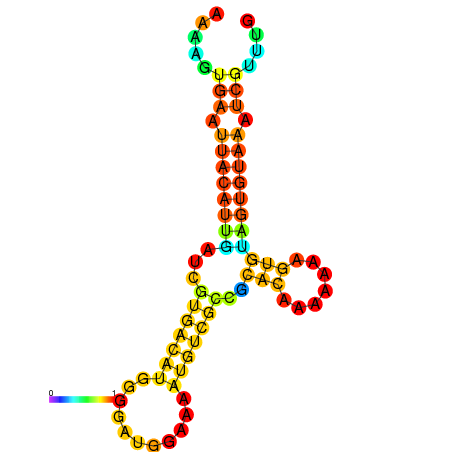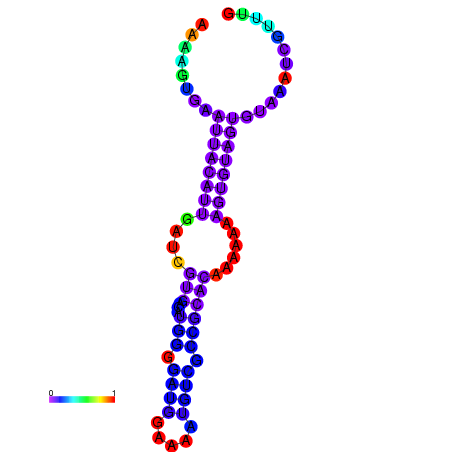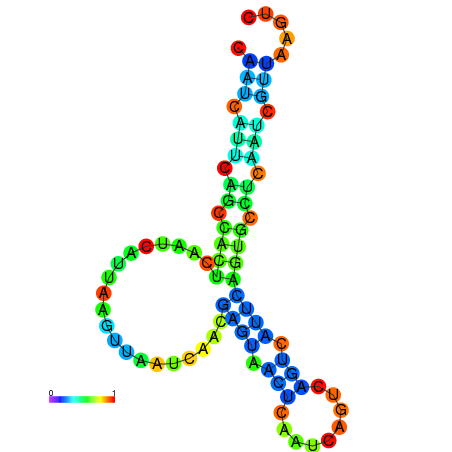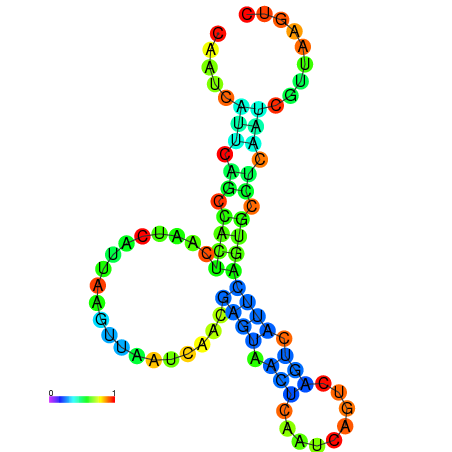| dm3 |
chr2R:20181738-20181869 + |
ATAAAAAGTATGTCACT------------------T-GGGG-GCGCTGCACT---------GGCATTGAAA-AGTGAA------TTACATTGATCGTGACATGGGAATGGAAAATGTCGCCG-----------------------------------------------------------------------------------------------------------------------------------------------AGCCATGTAAATCGTTTGGCCTACCCGTCTACCA-GCCAACC----AACCA |
| droSim1 |
chr2R:18643325-18643449 + |
ATAAAAAGTATGTCACT---------------TGGTG-GGG-GCGCTGCACT---------GGCATTGAAA-AGTGAA------TTACATTGATCGTGACATGGGGATGGAAAATGCCGCCG-----------------------------------------------------------------------------------------------------------------------------------------------AGCCATGTAAATCGATTG---------AAGGCC-C-GTGGCC----AACCA |
| droSec1 |
super_23:61187-61303 + |
ATAAAAAGTATGTCACT---------------TGGTG-GGG-GCGCTGCACT---------GGCATTGAAA-AGTGAA------TTACATTGATCGTGACATGGGGATGGAAAATGTCGCCG-----------------------------------------------------------------------------------------------------------------------------------------------AGCCATGTAAATCGATTG---------AAGGCC-C-GTG------------ |
| droYak2 |
chr2R:20147877-20148015 + |
ATAAAAAGTATGTCACT------------------G-GGGG-GCGCTGCACT---------GGCACTGAAA-AGTGAA------TTACATTGATCGTGACATGGGGATGCGAAATGTCGCCAAACGAAAAA----ATAAATCA----------------------------------------------------TAG------------------------------------------------------------------------TGTAACTCGTTTG---------AAGGCCACACCAACT----AGTAA |
| droEre2 |
scaffold_4845:21546843-21546980 + |
ATAAAAAGTATGTCACT---------------A----GGGGGCTTCTGCACT---------GGCATTGAAA-AGTGAA------TTACATTGATCGTGACATGGGGATGGAAAATGTCGCCGCACAAAAAA--------AGTG----------------------------------------------------TAG------------------------------------------------------------------------TGTAAATCGTTTG---------AAGGCC-C-ATGGCCAACCAACCG |
| droAna3 |
scaffold_13266:14229911-14230049 - |
AAAAAAAGTATGTCACA---------------C--TG-GGG-GAACTGCACT---------GACATTGAAAAACTGAA------TTACATTGGACGGTGGATGGG-------------------------------------------TGGACTGATGGACTGATGGACTGACAGCCA-------------------------------------------------------CGATCATC--------ACTAGCGTCGCAGTTAAGTTATGTAA-------------------------------------ACCG |
| dp4 |
chr3:18492568-18492741 - |
GTAAAAAGTATGTCAGAGAAGAGAAGAGAACACAGTGGGCG-CTGCTGCACT---------GACATTGAAA-AGTGAA------TTGCATTGATCATGACGGGGGGTTGA-------CGCTG-------------------------------------------------ACTGCCACGAACCGCTCAGAACAATAGTGTAAATGTGGAATAATGCTCG--------------------------------------------------TAGAAC-CCATTC---------G----CA-GTCCATG----GGCTG |
| droPer1 |
super_45:328754-328870 - |
GTGAAATGTATTTGAAA---------------------------------CG---------AGTAACGGAA-ATTGCA------TTATTTCAATTACAAA-----------------------------------TCCAATGAGAGGGAGAGGG-------ACAGGGA----C---------------------------------------------AGGG-------------------------------------------ACCCAGCACAA-----------CCCTAGGAGCCA-GCCAGCC----AGCCA |
| droWil1 |
scaffold_180949:4063354-4063494 - |
ATTGAGGGCGTGATTTA------------------TG-CCC-GATCTACCCC---------ATCATTTAAT-GTTAAAGACAATGCTAATTGGCCATTAAAATAGGCCG-----------CA------------------------ACAGACAC-------AAATAGT----T---------------------------------------------TGGGACCTA------AAATCAAA-------------------------------------------GCCAACCGACCACCA-GCCAACC----AGCCA |
| droVir3 |
scaffold_12875:7276052-7276211 + |
GG----------------------------------------GCGCTGCATTTGCCAGGCCAGAGTTAAAA-AGTGAA------TTGAATTGGCCGCTTGATTGA-------------------------------------------CAG-CCGACGGGCTGA-------------------------------------ATATGTGCAGCAATGCCAACAAATCGATGCCAAAGTCAGTGTCAGTCCCACGTAATCGACAATCAGTTATTTAC-----------------------A-TACGAGC----CACAA |
| droMoj3 |
scaffold_6473:1699609-1699741 - |
ATAATCTATTAGTTATT---------------TAATA-TAT-GT------------------------------CAATCATTCAGCCACTCAATCATTAA-----------------------------------GTTAATCAACGAGTAACTC-------AATCAGT----C---------------------------------------------AGTCATTCAGTGCCTCAATCGTT----------------------------------------AAGTCTTCCAATAAGTCA-CTCAATC----AGTCA |
| droGri2 |
scaffold_15203:4626832-4626941 - |
GCCCATGCCA--------------------------------------------------------TAAAA-AGAGTT------TTGAACTAATCTTAAGCAGGCAGCGCAAATCGCTATAAATCAG-CTTTCTATTCTT--------------------------------------------------------TG------------------------------------------------------------------------TT---------TTGCCCCACTGTGCACCA-GCCAGCC----AGCCA |