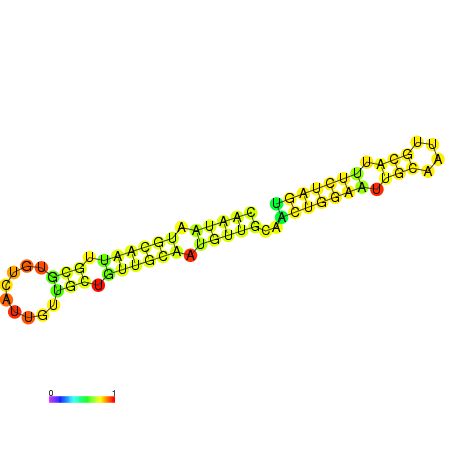
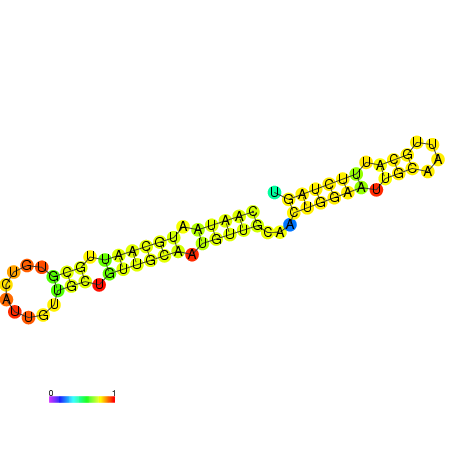
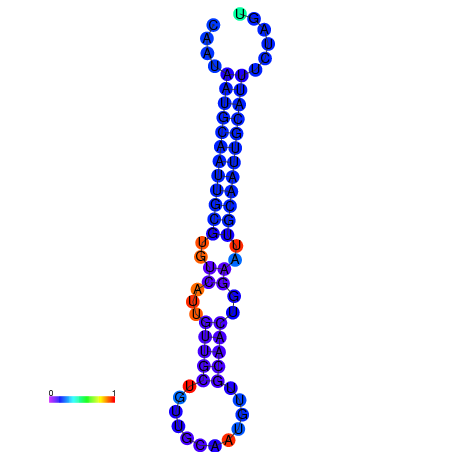
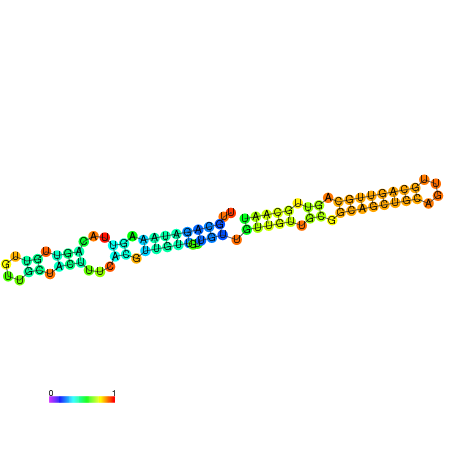
| dm3 |
chr2R:14607658-14607807 + |
TTTT-GAGTCGAAAGGACC--A----------------ACT-TTGCTGGCATTCGGTGGCCAA--------------TAATGCAATTGCATGTC-------------------------------------------------------CTTGTTGCTGTTGCAAT----GTTG-------CAGCTGGAATTGCAATTGCATTTCTAGTTGC------------------CGCTGCTG---------CTGCTCGAGTGGCAATTA--GCTGGGTGTCTC |
| droSim1 |
chr2R:13317686-13317836 + |
TTTTCGAGTCGAAAGGACC--A----------------ACT-TTGCTGGCATTCGCTGGCCAA--------------TAATGCAATTGCGTGTC-------------------------------------------------------CTTGTTGCTGTTGCAAT----GTTG-------CAGCTGGAATTGCAATTGCATTTCTAGTTGC------------------CGCTGCTG---------CTGCTCGAGTGGCAATTA--GCTGGGTGTCTC |
| droSec1 |
super_1:12115359-12115508 + |
TTTC-GAATCGAAAGGACC--A----------------ACT-TTGCTGGCATTCGCTGGCCAA--------------TAATGCAATTGCATGTC-------------------------------------------------------CTTGTTGCAGTTGCAAT----GTTG-------CAGCTGGAATTGCAATTGCATTTCTAGTTGC------------------CGCTGCTG---------CTGCTCGAGTGGCAATTA--GCTGGGTGTCTC |
| droYak2 |
chr2R:14591968-14592144 - |
TTTTCGGGTCGAAAGGACC--A----------------ACTTTTGTTGGCATTCGCTGGCCAA--------------TAATGCAATTGCGTGTC-------------------------------------------------------CTTGTTGCTGTTGCAAT----GTTGCAACTTGCAACTGGAATTGCAATTGCATTTCTAGTTGCTTCTCCTCTTTTTCCTGCTGCTCCTA---------CTCCTCGAGTGGCAATTA--GCTGGGTGTCTC |
| droEre2 |
scaffold_4845:8798992-8799150 + |
TTTC-GCGTCGAAAGGACC--A----------------ACT-TTGCTGGCACTCGCTGGCCAA--------------TAATGCAATTGCGTGTC-------------------------------------------------------ATTGTTGCTGTTGCAAT----GTTG-------CAACTGGAATTGCAATTGCATTTCTAGTTGCTCCTCA---------TGCTGCTGCTG---------CTCCTCGAGTGGCAATTA--GCTGGGTGTCTC |
| droAna3 |
scaffold_13334:1368045-1368196 + |
TTTT-GAGCTGCAG-----------------------------------------------------------------------------------------------TTGCAGATAAAGTTACAGTTGTTGTTGCTACTTTCACGTTGTTGTTGTTGTTGT--T----GCGG-------CAGCTGCAGTTGCAGTTGCAGTTGCAATTGT------------------TGCTGCAGCTGTTGGCCTAATGAGAGTGGCCAGCAATGCGGCACGAATC |
| dp4 |
chrXL_group3a:862470-862596 - |
---------------------A----------------ACT-CTGCTGCCAGTAGTTGGCCAT--------------TGAGAAAATTGTGTGCCCTGGAATTTAATTCATTGCAAACAAA-------TTGCT-----------------GCTGCTGCTGCTGC----------------------------------TGCTGCTGTTGTTGC------------------TGCTGCTG---------CCGTTGCAGTGGCAATT--------------- |
| droPer1 |
super_15:2034074-2034206 + |
---------------------A----------------ACT-CTGCTGCCAGTAGTTGGCCAT--------------TGAGAAAATTGTGTGCCCTGGAATTTAATTCATTGCAAACAAA-------T-----------------------TGCTGCTG----------------------CTGCCGCTGCTGCTGTTGTTGTTGTGGTTGC------------------TGCTGCTG---------CCGTTGCAGTGGCAATT--------------- |
| droWil1 |
scaffold_181096:6674642-6674780 + |
CTTG-GTGCCGTAT----CAAAGCTGGCATATAGTTCAACT-TCT------T-CATCGG----TGGCATCA------CGTTGCAGTTGT-TGTT-------------------------------------------------------GTTGTTGTTGTTGC-------------------------------TGTTGCAGCTGTTGTTGTTGCTGTTGTTGTTGCTGCAGCTGCTG---------CTGCTCAAGTG--------------------- |
| droVir3 |
scaffold_12822:1548557-1548661 - |
TGTT-------------------------------------------------------------------------------------------------------------------------------------------------GCTGCTGCTGTTGCAATTGTTGCTG-------CAGCTGTTGTTGCTGTTGCAGCTGCAGCTGTAACTGT---------TGCTGC---TG------CTGGGCATAGAGCTGCTCCTG--GT--AGTGCTTG |
| droMoj3 |
scaffold_6540:8945968-8946087 + |
AGCC------AGCATCGAC--A----------------ACT-TGGCTGCAATTCAGGT-------------TGCAGCTACTGCTGTTGCCTGTT------------------------------------------------G------CCTGTTGCCGTTGCC------GTTG-------CTGCTGCAGTTGCAGTTGCAGTTGTAGTTAC------------------CGTTGCAA---------TTGC---------------------------- |
| droGri2 |
scaffold_15245:16288597-16288735 + |
TGTT-G-----------TC--A----------------GCG-TTGTCGTTG--TGCTTGTTGA--------------TTTTCCAGTTGCT-----------------------------------------------------------GCTGTTGCTGTTGCAGTTGCAGTTG-------CAGCTGTTGCTGCTGTTGCAGTTGCAGTTGTTGCTGC---------TGCTGC---TG------TTGTTGTTGGTGTT---------GTCGGTGGTGTC |