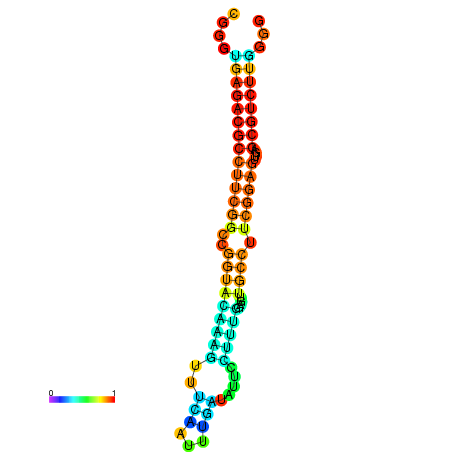
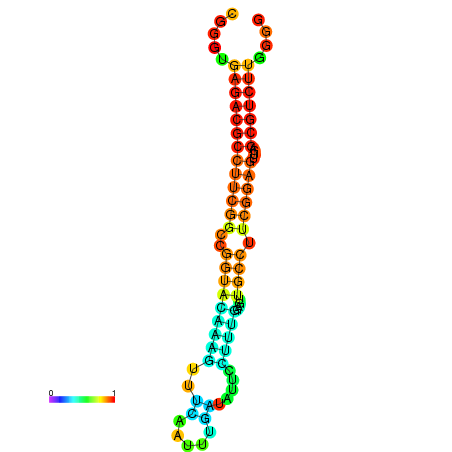
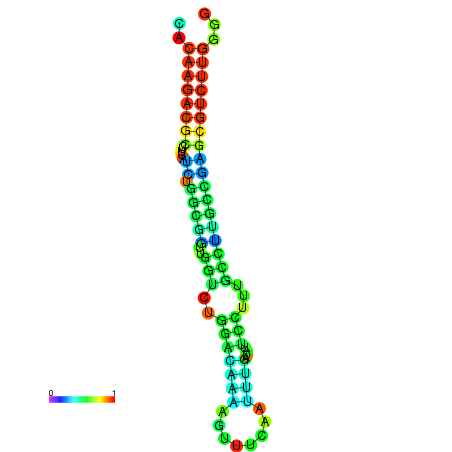
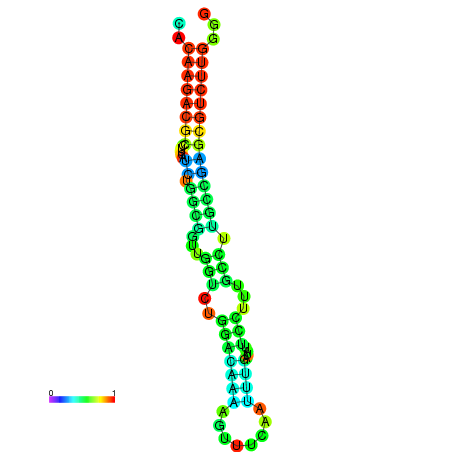
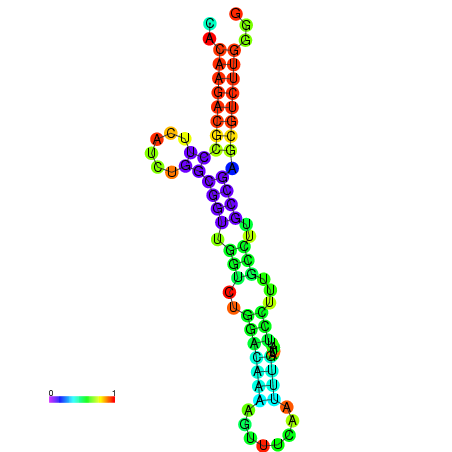
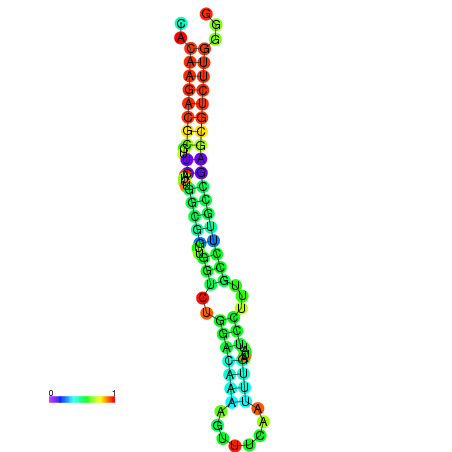
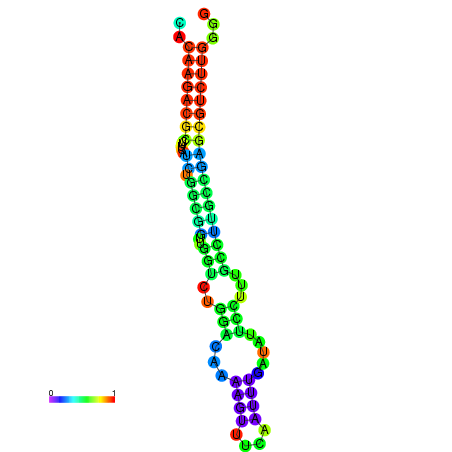
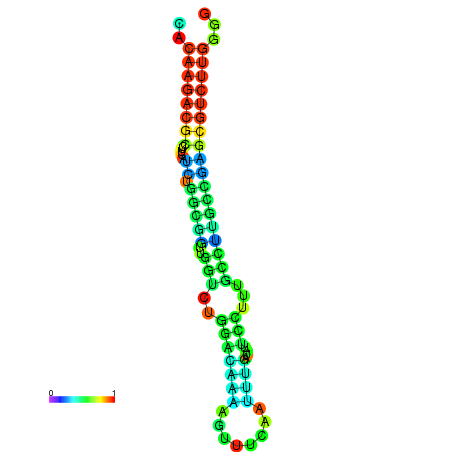
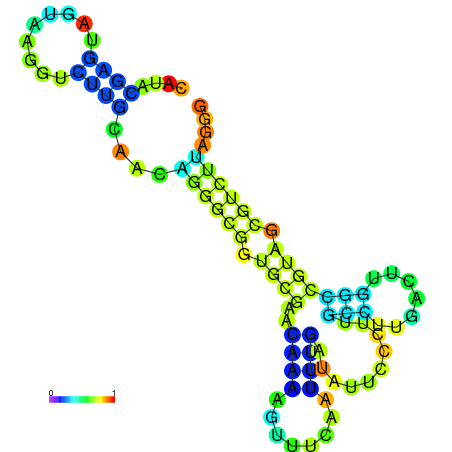
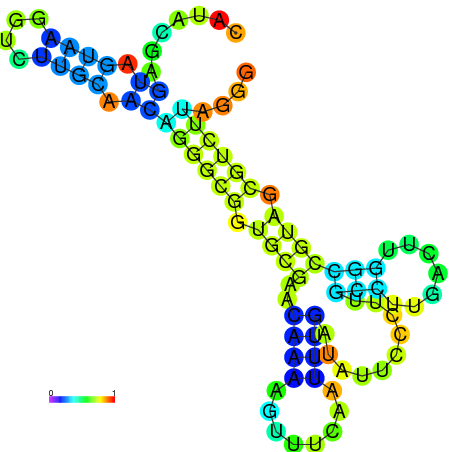
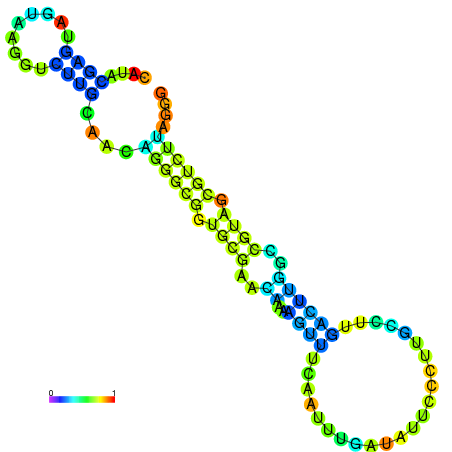
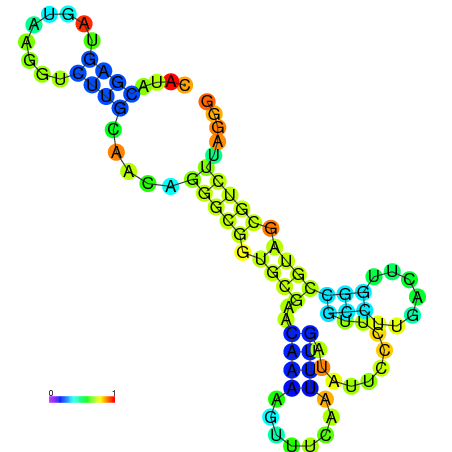
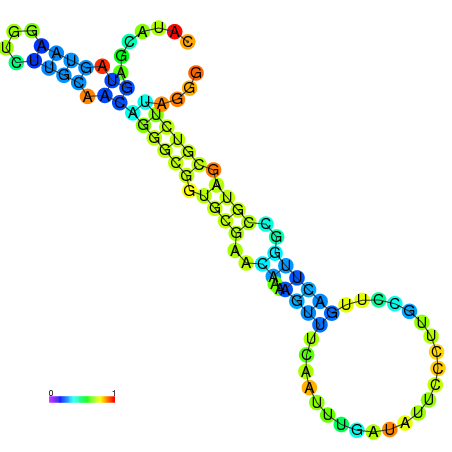
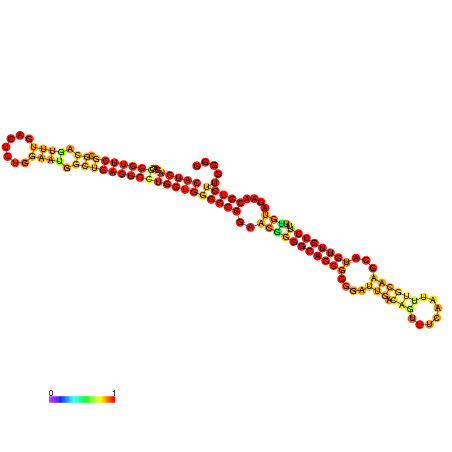
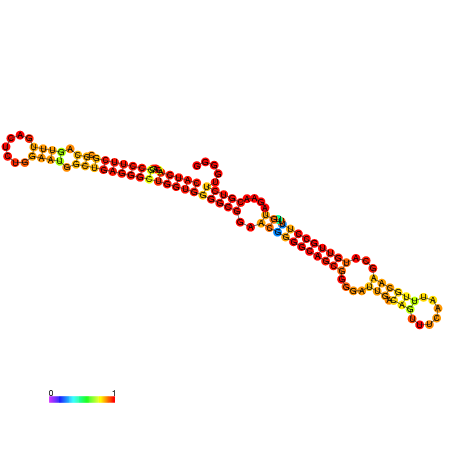
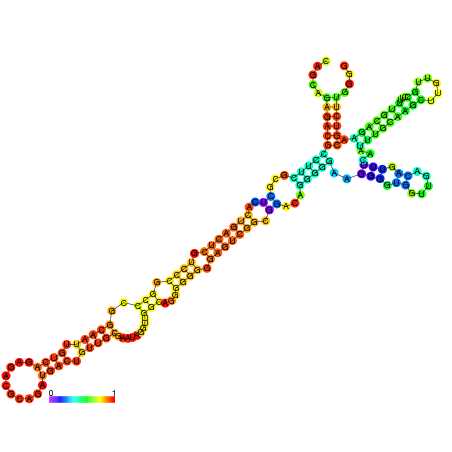
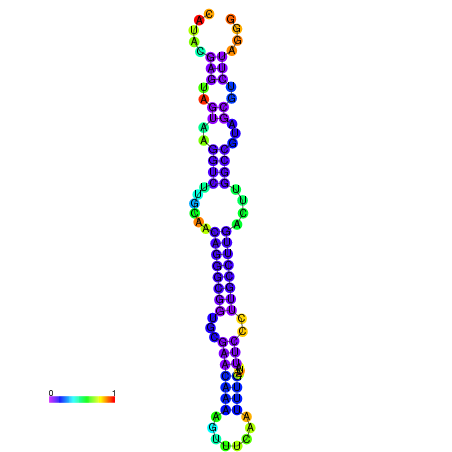| dm3 |
chr2R:3902738-3902863 + |
GGTTATC-------------------------------------CCTGC-CACATGGCCACGTGCCCG-CAAGACGCCTTCGGCCGGCG-------------------------------------------------------------------------------------------------------CAAAGTTTCAATTTGATATTCCTTGGC-CG-----GTCGAG--CAGCGTCTTGGGG-TTGTTCG--------TGGATTAAGTTCGATTC--------------------------CTAT |
| droSim1 |
chr2R_random:1106077-1106202 + |
GGATATC-------------------------------------CCTGC-CACATGGCCACGTGCCTG-CAAGACGCCTTCGGCCGGCG-------------------------------------------------------------------------------------------------------CAAAGTTTCAATTTGATATTCCTTGGC-CG-----GTCGAG--CAGCGTCTTGGGG-TTGTTCC--------TGGATTAAGTTCGATTC--------------------------CTAT |
| droSec1 |
super_1:1540674-1540799 + |
GGATATC-------------------------------------CCTGC-CACATGGCCACGTGCCCG-CAAGACGCCTTCGGCCGGCG-------------------------------------------------------------------------------------------------------CAAAGTTTCAATTTGATATTCCTTGGC-CG-----GTCGAG--CAGCGTCTTGGGG-TTGTTCC--------TGGATTAAGTTCGATTC--------------------------CTAT |
| droYak2 |
chr2L:16559915-16560042 + |
GGATATC-------------------------------------CCTGC-CACATGGCCACGTGCCTG-CAAGACGCCTTCGACCGGCG-------------------------------------------------------------------------------------------------------CAAAGTTTCAATTTGATATTCCTTGGC-CG-----AACGAACGGAGCGTCTTGGGG-TTGTTTC--------GGGATTACGTTCGATTC--------------------------CGAT |
| droEre2 |
scaffold_4929:18723379-18723500 - |
GGATA-C-------------------------------------CCAGC-CACATGGCCACGTGCCTG-CAAGACGCCTTCGGCCGGCG-------------------------------------------------------------------------------------------------------CAAAGTTTCAATTTGATATTCCTTGGC-CG-----GTC-----GAGCGTCTTGGGG-TTGTTTC--------GGGATTGAGTTCGATTC--------------------------CGAT |
| droAna3 |
scaffold_13266:16222514-16222662 + |
GGAGATC-------------------------------------CTTG--CGTGTGGCCACGTGCCGGGTGAGACGCCTTCGGCCGGTA-------------------------------------------------------------------------------------------------------CAAAGTTTCAATTTGATATTCCTTTGGAGTTGCCTTCGGAGTTGAGCGTCTTGGGG-TTGTTCC--GGGCCATGGATTAAGTTG-----AGAAA-ATG------CAG----A-CTATGT |
| dp4 |
chr3:5536552-5536721 + |
GGTTG-C---------G------------CTGCTGCTTATGGTG----CTGGTGTTGCCACGTGCC-A-CAAGACGCCTTCATCTGGCGGTT------------------------------------------------------------------------------------------GGT-CTGGACAAAAGTTTCAATTTGATATTCCTTTGC-CT-----T--GCC--GAGCGTCTTGGGG-TTGTTTC--AGGGCATGGATTAAGTTT---TCAGGCAACTGGAGCAGCA------TCCGTGC |
| droPer1 |
super_2:5730715-5730884 + |
GGTTG-C---------G------------CTGCTGCTTATGGTG----CTGGTGTTGCCACGTGCC-A-CAAGACGCCTTCATCTGGCGGTT------------------------------------------------------------------------------------------GGT-CTGGACAAAAGTTTCAATTTGATATTCCTTTGC-CT-----T--GCC--GAGCGTCTTGGGG-TTGTTTC--AGGGCATGGATTAAGTTT---TCAGGCAACTGGAGCAGCA------TCCGTGC |
| droWil1 |
scaffold_181141:2380508-2380690 - |
ACCCATTTCTGGTCTCG------------GTCTCAATTTTTTTA----CTGGTGTGGCCACGTGCCA--TACGAG-----------------------------------------------------------------------------------TA-GTAAGGTCTTGCA-ACAGGGCGGT-GCGAACAAAAGTTTCAATTTGATATTCCCTTGC-CTTGACTTGGCCG--TAGCGTCTTAGGG-ATGTTTCC-GAAGCAAGGATTAAGTTT---TCAAA----G------GCAG------CTGTGC |
| droVir3 |
scaffold_12875:14800808-14801017 + |
GCTTCTTTG------------------------TGTCCATGGCCCTTGC-CGTGTGGCCACGTGCCA--TCAGACGCCTTCGCGCAGT---TTGACTC--------------------------------------------TGGAATGGCTGAGGGCTG-GTGGGG---CGGA-ACGGGGCAGCGGGGATTGACAGTTTCAATTTGCAAGCATGTTGC-CT-----TTTGTA--GAACGTCTTGGGGGTTGTTCGCCAGCTGACGGATTAAGTTT---TGAGGGC-A----------GCCAGATGCTGCT |
| droMoj3 |
scaffold_6496:7113415-7113501 - |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACAGTTTCAATTTGCAAGCTTGTTGC-CT-----TTTACG--GAACGTCTTGGGG-TGTCTGA--------GGGATTAAGTTTAAGTCTGGGC-AT---------GCCAG--C--TGC |
| droGri2 |
scaffold_15245:14811478-14811742 + |
GTTTCTTTGTG-TCCAGACCCTTGCCGCCTTGCCGC--------CTTGC-CGTGTGGCCACGTGCCAGCAGAGACGCCTTCGCGCTCA---CTGACTCGTCCCGGCCCGGCAATTGTCAGAGACGCAGATGACTGTTGCGAATAGGTTGGCAGGGGGGGGAGT-CGG---CGGACAGGGGGAAGGGGTGGTTGACAGTTTCAATTTGCAAGCTTGTTGC-CT-----TTTGCA--GAACGTCTTGGGG-TTGTTTC--AGCTGGCGGATTAAGGTT---TCAGACAG-T-------CAGCC-------AGC |