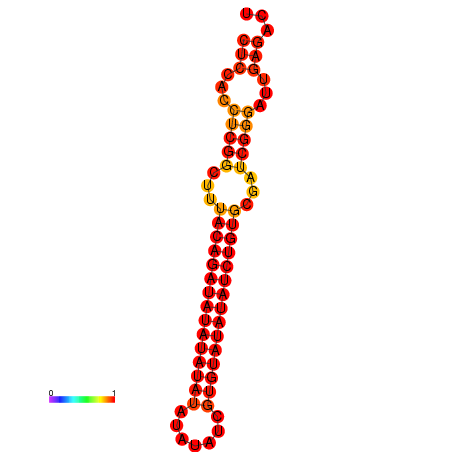
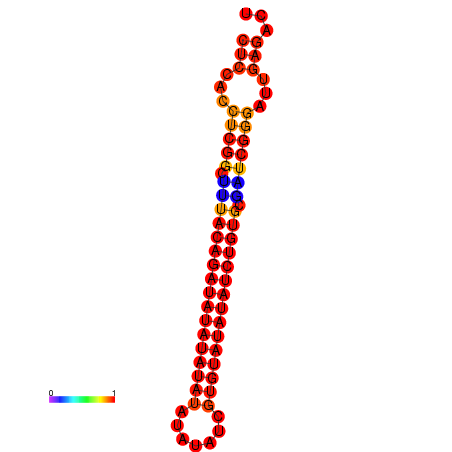
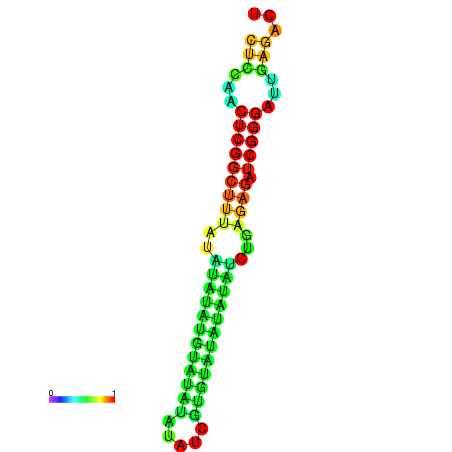
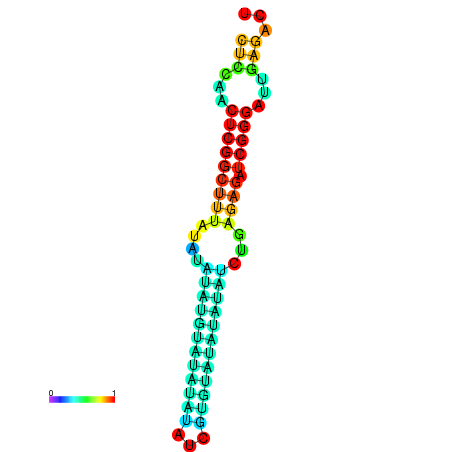
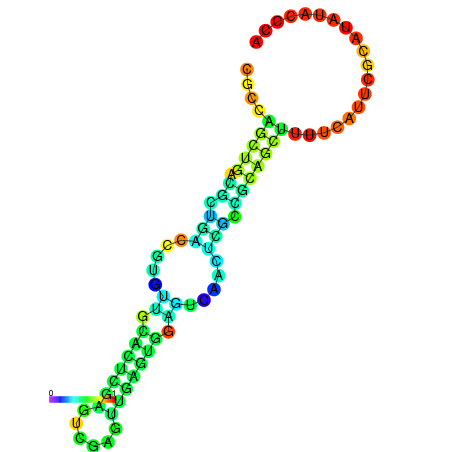
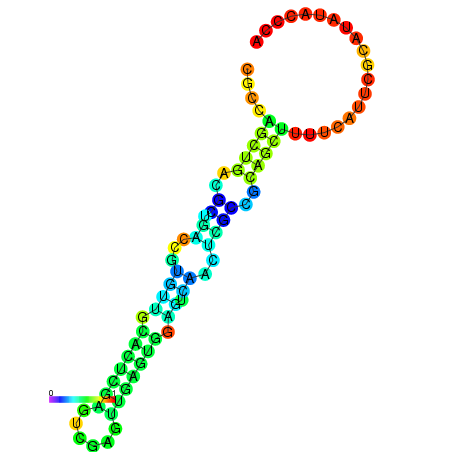
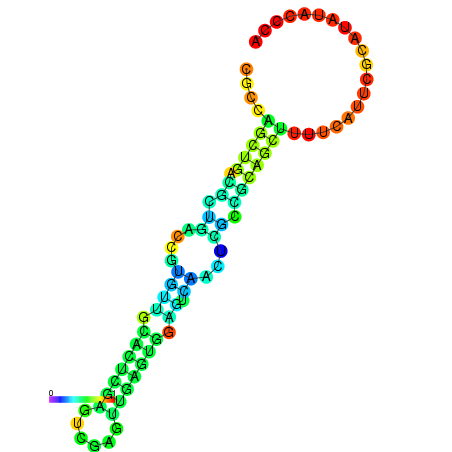
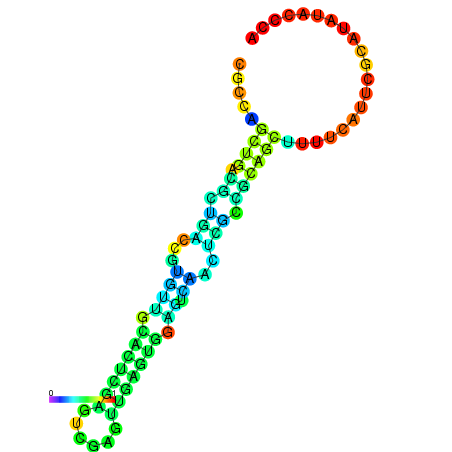| dm3 |
chr2L:3636951-3637088 - |
CACTTGCATGGCATAAATCAAAAGCGCCGTAGGCCT-CCACCTCG--------------GC---------------------------------------------------------------------------------------TTTACAGATATATATATATATATCGTGTATATATCTGTGCGATCGGG-A-TTGAGACTGAGGTTGGT-----CCGAG-TACTTTTGTGGCCA--TTTACACC |
| droSim1 |
chr2L:3596396-3596533 - |
CACTTGCATGGCATAAATCAAAAGCGCCGTAGGCCT-CCAACTCG--------------GC---------------------------------------------------------------------------------------TTTATATATATGTATATATATCGTGTATATATATCTGAGAGATCGGG-A-TTGAGACTGAGGTTGGT-----CCGAG-TACTTTTGTGGCCA--TTTACACC |
| droSec1 |
super_5:1737908-1738045 - |
CACTTGCATGGCATAAATCAAAAGCGCCGTAGGCCT-CCACCTCG--------------GC---------------------------------------------------------------------------------------TTTATATATATGTATATATATCGTGTATATATATCTGAGAGATCGGG-A-TTGAGACTGAGGTTGGT-----CCGAG-TACTTTTGTGGCCA--TTTACACC |
| droYak2 |
chr2L:3630931-3631061 - |
CACTTGCATGGCATAAATCAAAAGCGCCGTAGGCCT-CCAACTCG--------------AC---------------------------------------------------------------------------------------TATATATGTATATATATA-------TATATATATCTGTGAGATCGGG-A-TTGAGACTGAGGTTGGT-----CTCAG-TACTTTTGTGGCCA--TTTACACC |
| droEre2 |
scaffold_4929:3675838-3675960 - |
CACTTGCATGGCATAAATCAAAAGCGCCGTAGGCCT-CCACCTCA--------------GC---------------------------------------------------------------------------------------TATATATATATGT---------------GTGTATCTGTGGGATCGGG-A-TTGAGACTGAGGTTGGT-----CTTAG-TACTTTTGTGGCCA--TTTGCACC |
| droAna3 |
scaffold_12916:5691742-5691836 - |
CACTTGCAAGGCATAAATCAAAAGCGCCGTAGGCCT-CCGCCACT------------------------------------------------------------------------------------------------------------------------------------------------ATGAGT-C-TGGAGTCTGGTGATGGT-----GGAGT-GCCTTTTGTGGCCA--TTGGCACC |
| dp4 |
chr4_group3:5095808-5095886 + |
CACTTGCGTGGCATAAATCACAAGCGCCGCAGGCCT-GAGACTT-TGGAGTTTTTTGTGGCACAG-------------------------------------------------------------------------------------TGTGTGT--GTG----------------------------------------------------------------------------------TGTGTG |
| droPer1 |
super_1:6587322-6587402 + |
CACTTGCGTGGCATAAATCACAAGCGCCGCAGGCCT-GAGACTT-TGGAGTTTTTTGTGGCACAG-------------------------------------------------------------------------------------TGTGTGTATGTG----------------------------------------------------------------------------------TGTGTG |
| droWil1 |
scaffold_180708:9512363-9512488 + |
CACTTGCATGGCATAAATCAAAAGCGCCGTAGGCCTTCCACCTCT---------ATAT-------AGTATAATGTGTTCCACATACTCGAC---------------------------------------------------------------------------------------------------TTGGG-AGTTGAATTCGAAGTTG-T-----CCATG-T-CTTTTGTGGCCAATTTTACACC |
| droVir3 |
scaffold_12963:15108661-15108745 + |
CACTCGCGTGGCATAAATCAAATGCGTTGCAGGCCG-CCGGCGCT-------CCACGTT-----------------------GCACTCGACTC--------------------------------------------------------------CT-------------------------------------------------------------------C-TA--CTTGCGGCTA--GCAGCTCC |
| droMoj3 |
scaffold_6500:18077427-18077576 + |
CACTTGTGTGGCATAAATCAAATGCGCAGCAGGCCG-CCAGCTGA--------------------CGCTGACCGTGTT----GCACTCGAGTCGAG------------------------TTGAGTGGA---GTCAACTCGCCGCAGCTTTTCATTCGCATATACCCA-----------------------------------------GCCAAGCCAAG-----TGGTCTTTGGGGCCA--TTTACACG |
| droGri2 |
scaffold_15126:4287105-4287265 - |
CACTTGCATGGCATAAATCAAATGCGCCGCGGGCCT-TTGCTACG-----------------------------TGCCACATGCACTCGACTCTCGCCTCTCAACACGACTGACCGAACATTGGCTGGACAAGCCAAGCTGCAG------------------------------------------CAGATCGGGGCATTCCACTCCAAG---------------TGGTCTTTGAGGCCA--TTTGCACG |