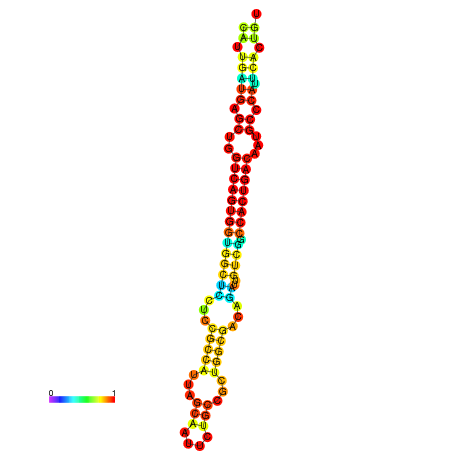
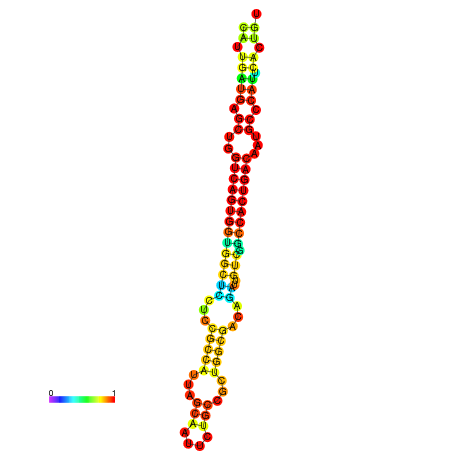
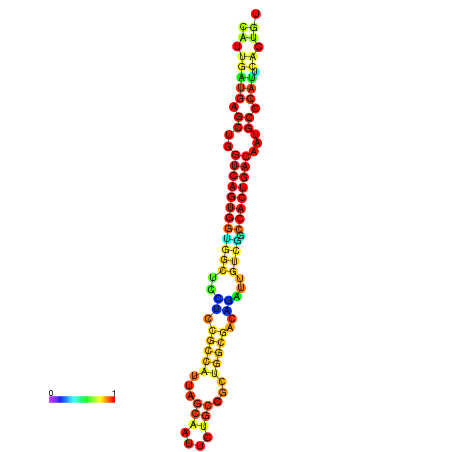
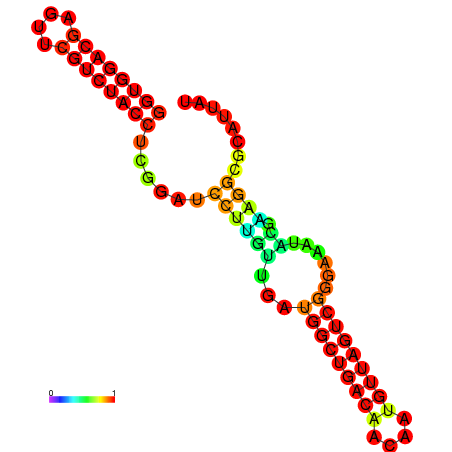
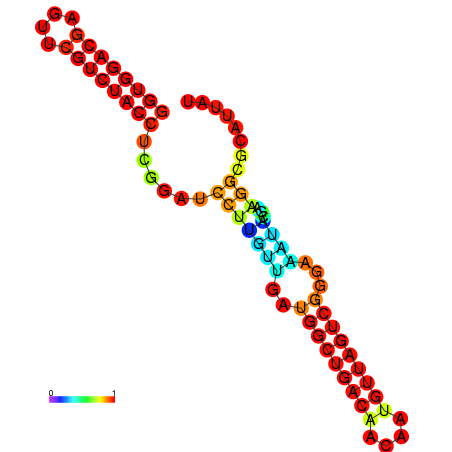
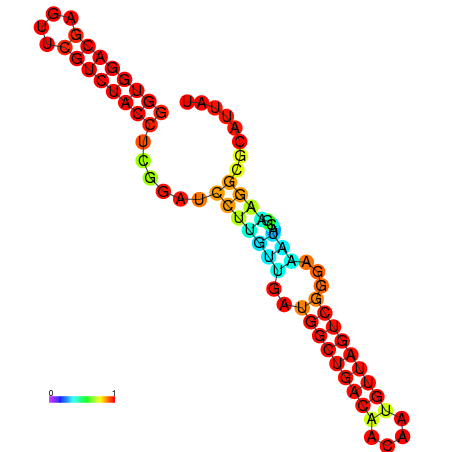
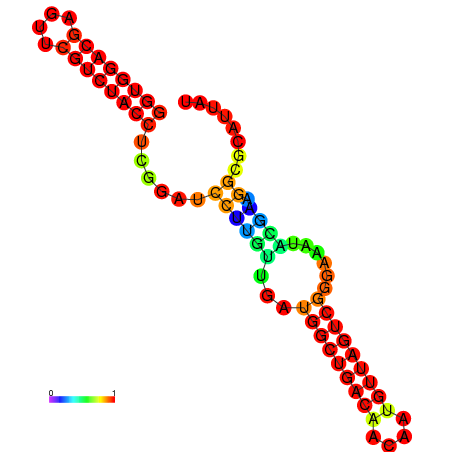
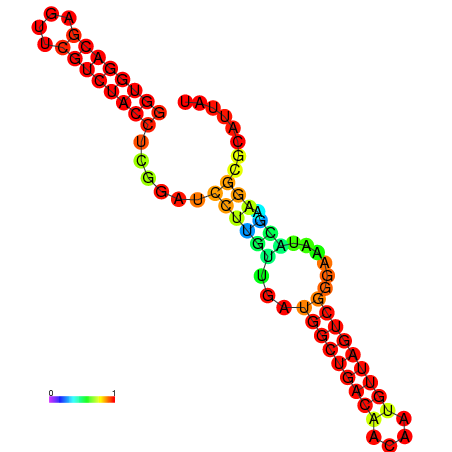
| dm3 |
chr2L:3463274-3463415 - |
GGAAG-GGCCAACTTCT---CGGCCCT--------CGACACGGTGGATGAGCTTGTCAGTGGCGGATC------CAGGAGCAATTCTGCCGC-------TGGCGACAG--ATCATCGGCCA--CTGACAATGC----------------------CCATTCACTGTTCGAGGAGATCATGTCGG--------GCTCGGATG |
| droSim1 |
wop21h10.b1:281-423 + |
GGAAG-GGCCAATTTCT---CGGCTCT--------CGACACGGTTGATGAGCTTGTCAGTGGCGGATC------CAGGAGCAATTCTGCCGC-------TGGCGACAG--ATCATCGGCCA--CTGACAATGC----------------------CCAGTCACTGTTCGAGGAGATTATGTCGG--------GCTCGGATG |
| droSec1 |
super_129:25371-25512 - |
GGAAG-GGCCAACTTCT---CGGCCCT--------CGACACGGTTGATGAGCTTGTCAGTGGCGGATC------CAGGAGCAATTCTGCCGC-------TGGCGACAG--ATCATCGGCCA--CTGACAATGC----------------------CCAGTCACTGTTCGAGGAGATCATGTCGG--------GCTCGGATG |
| droYak2 |
chrU:14964015-14964156 + |
GGAAG-GGCCAACTTCT---CGGGCCT--------CGACACGGTGGATGAGCTTGTCAGTGGCGGATC------CAGGAGCAATTCTGCCGC-------TGGCGACAG--ATCATCGGCCA--CTGACAATGC----------------------CCACTCACTGTTCGAGGAGATCATGTCGG--------GCTCGGATG |
| droEre2 |
scaffold_4929:3498112-3498253 - |
GGAAG-GGCCAACTTCA---CGGGCCT--------CGACACGGTGGATGAGCTTGTCAGTGGTGGATC------CAGGAGCAATTCTGCCGC-------TGGCGACAG--ATCATCGGCCA--CTGACAATGC----------------------CCACTCGCTGTTCGAGGAGATCATGTCGG--------GCTCGGATG |
| droAna3 |
scaffold_12943:2497935-2498076 + |
GGCAG-AGCCAACTTCG---GAGCCCT--------TGAAACGGTCGACGAGCTTGTCAGTGGCGGCTC------CATAAAGAATCCTGCCGC-------TGGCGACAG--ACAGTCGGCCA--CTGACAACGC----------------------GCACTCGCTGTTCGAGGAAATAATGTCTG--------GCTCCGACG |
| dp4 |
chr4_group3:936699-936854 - |
GGAGT-GCTCGTCTACG---TGGTTTTGCCCCACGCAATACCATTGATGAGCTGGTCAGTGGTGGCTCCTCCGCCATTAGCAATTCTGCCGC-------TGGCGACAG--ATTGTCGGCCA--CTGACAATGC----------------------CCATTCACTGTTCGAGGAGATCATGTCGG--------GCTCTGATG |
| droPer1 |
super_1:2415168-2415323 - |
GGAGT-GCTCGTCTACG---TGGTTTTGCCCCACGCAATACCATTGATGAGCTGGTCAGTGGTGGCTCCTCCGCCATTAGCAATTCTGCCGC-------TGGCGACAG--ATTGTCGGCCA--CTGACAATGC----------------------CCATTCACTGTTCGAGGAGATCATGTCGG--------GCTCTGATG |
| droWil1 |
scaffold_189765:461-602 - |
GTAAC-GATAGACGGAG---ATACCTT--------CGAGGTGGTGGACGAGTTCGTCTACCTCGGATC------CTTGTTGATGGCTGACAA-------CAATG----------TTAGTCG--GGAAATACGA----------------------AGGCGCATTATCAGCGGAAGTCGTGCCTACTATTGGCTCCAGAAGA |
| droVir3 |
scaffold_12963:15952027-15952160 - |
ATTAGATGCCGATTGCGGTTCGTCCAT--------CAAGTCTGCGGCTGAGCTTGTCAGCGCCGT---------------CAATGCTGCCGC-------TGGCGACAG----CCTCGGC-G--CTGACAAGGA----------------------GCATGCCATGTTTGAGTATTTCCTCAACG--------AAGATGACG |
| droMoj3 |
scaffold_6473:12510219-12510341 + |
GGTTT-G----------------------------------------TGTGTGTGTGTGTGGTGGAACTTC---AATGCGAACCT------CTTGCCACAATCGAGTGCAATTAACTGCCA--TGATATATGGCCAATCCCA-------------------TATGCAGACGGCGACCGAGTCGAA-------GTTCGGCTG |
| droGri2 |
scaffold_14853:4216060-4216150 - |
------------------------------------------------------------------------------------TCGACCGC-------TGGCCACAA--ATGGCCGGACAGACAGACA-------GACAGACAGACAGACAGACAGACTCACTGACATAATTGATCATATC----------GCTTATACG |