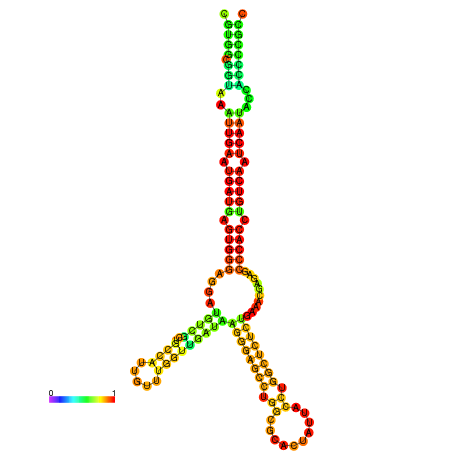
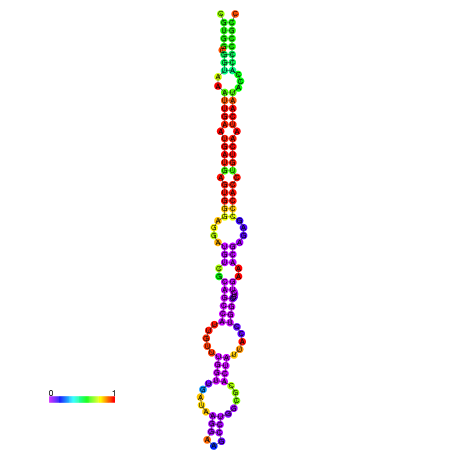
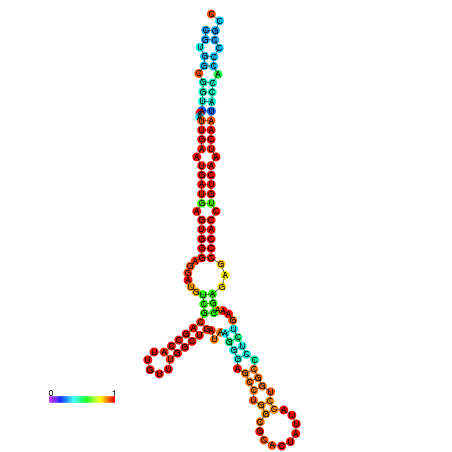
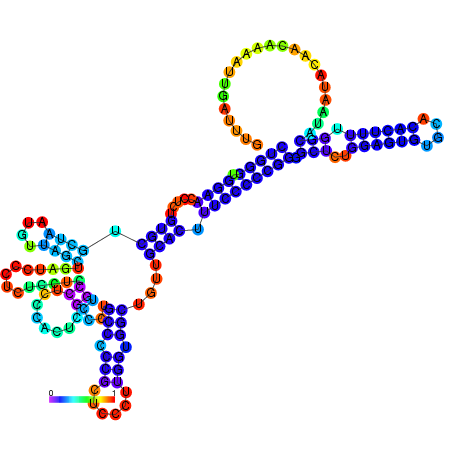
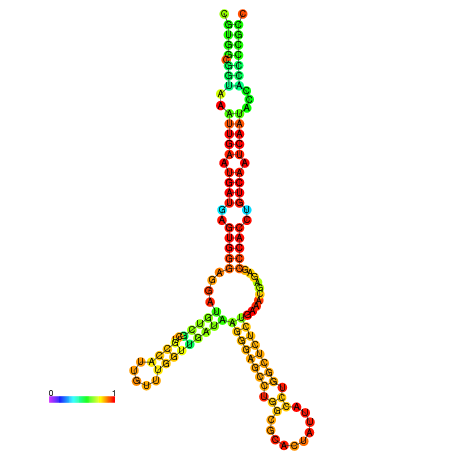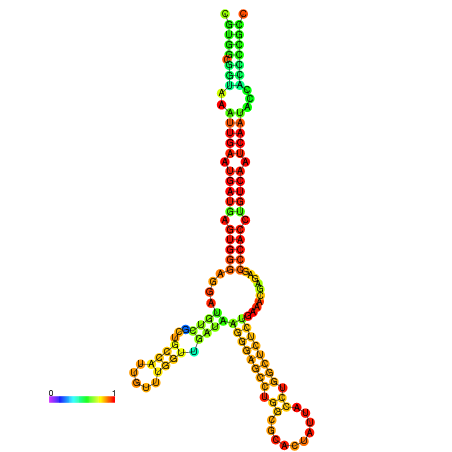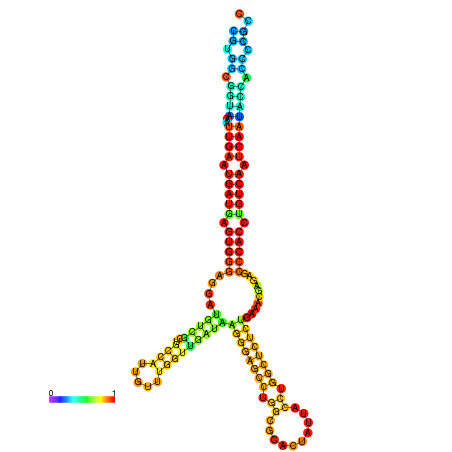| dm3 |
chr3R:17624220-17624408 - |
TATATATATGTTTTTGTATGGATTTTTCGGCGTGGCGGTAA------------ATTGAATGATGAGTGG-GAGG--ATGTCGCTGCCATTG----------TTTGG---TTGATA--------------------------------------AGGGAGCCTGGCGCACTATTACCTGGCTCTCTGAA-------------------------------------------A-CGAGAGC--------CCACCTGTCA--------ATCAATACCACCCCG-------CCGA--TGCGACATCATCATAATAATCATCGCCTTT |
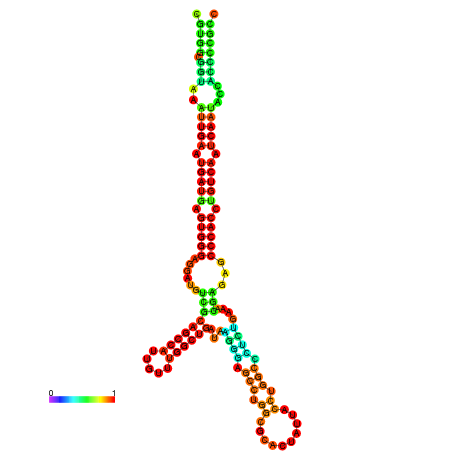
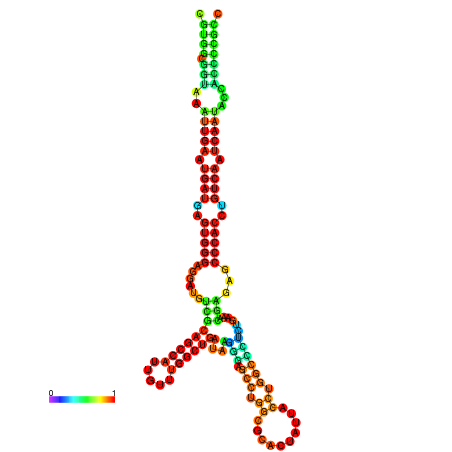
| droSim1 |
chr3R:17429869-17430050 - |
TGTTTTTG--------TATGGATTTTTCGGCGTGGCGGTAA------------ATTGAATGATGAGTGG-GAGG--ATGTCGCAGCCATTG----------TTTGG---TTGATA--------------------------------------AGGGAGCCTGGCGCACTATTACCTGGCTCTCTGAA-------------------------------------------A-CGAGAGC--------CCACCTGTAA--------ATCAATACCACCCCG-------CCGA--AGCGACATCATCATAATAATCATCGCCTGC |
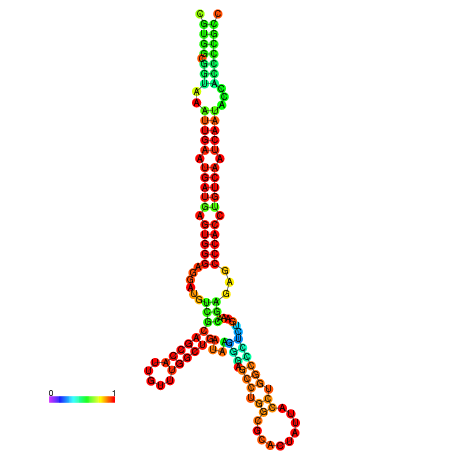
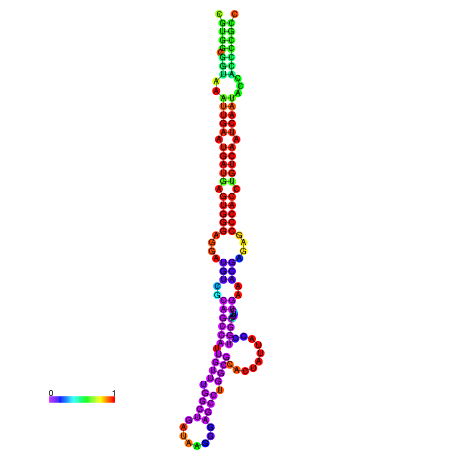
| droSec1 |
super_0:17996591-17996767 - |
TATATATATGTTTTTATATGGATTTTTCGGCGTGGCGGTAA------------ATTGAATGATGAGTGG-GAGG--ATGTCGCAGCCATTG----------TTTGG---TTGATA--------------------------------------AGGAAGCCTGGCGCACTATTACCTGGCTCTCTGAA-------------------------------------------A-CGAGAGC--------CCACCTGTCA--------ATCAATACCACCCCG-------CCGA--AGCGACA------------TCATCGCCTTT |
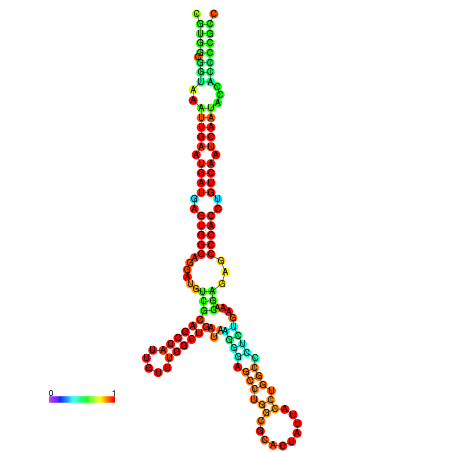
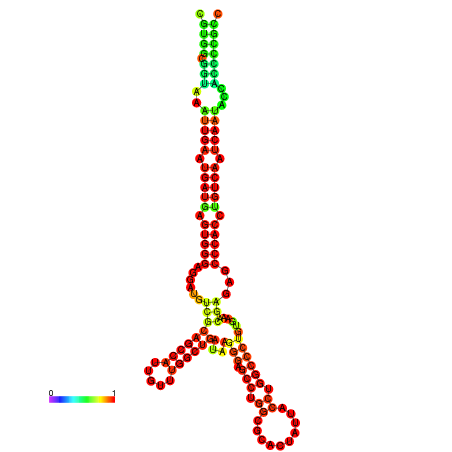
| droYak2 |
chr3R:18444204-18444378 - |
G---------------TATGGATTTTTCGGCGTGGCGGTAA------------ATTGAATGATGAGTGG-GAGG--ATGTCGCAGCCATTG----------TTTGG---CTGATA--------------------------------------AGGGAGCCTGGCGCACTATTACCTGGCCCTCTGAA-------------------------------------------A-CGAGAGC--------CCACCTGTCA--------ATCAATACCACCCCG-------CCGA--ACCGACATCATCATAATAATCATCGCCAGC |
| droEre2 |
scaffold_4820:10462895-10463063 + |
TGTAT--------------GAATTTTTCGGCGTGGCGGTAA------------ATTGAATGATGAGTGG-GAGG--ATGTCGCAGCCATTG----------TTTGG---CTGATA--------------------------------------AGGGAGCCTGGCGCACTATTACCTGGCCCTGTGAA-------------------------------------------A-CGAGAGC--------CCACCTGTCA--------ATCAATACCACCCCG-------CCGA--ACCGACATCATCATAGTAATCAT------- |
| droAna3 |
scaffold_13417:705028-705198 + |
TGTATATGTGTGGGTGTAGAGGTGTGT--------AGGTGT------------GCAGGTGTACGGTTGT-GTGT--GTGTGGCATTCGTTGCT-ACTTATGTTTAA-GCTTCGTA--------------------------------------TTTAAGCTTTTTGCACGATTTCCT----CTAACAA-------------------------------------------A-TGGATTT--------TCACTTGTT------------------GCCCAGCCAG---ACG---AACGAAGCCATCACAAAAAT---------- |
| dp4 |
chr4_group3:1192892-1193069 - |
TATATGGCCTATCCAGCAGGAATTTATCCAGCGGATGGTAG------------CTAGAGATGC-AGTTG-GAGG--GGG---GCACCATAG----------TTTGG---AGTAGC--------------------------------------CAAA---------------TGCCAGGCGATAAGGC-------------------------------ATGAAAAATACAA-CTAATAA--------ACATAAACCAGTCTTCTTTTAAAA--------G-------CTGC------TCATCATCATCATCATCATGATCATC |
| droPer1 |
super_7:2291330-2291534 - |
CATAAAAGGGTCTGAGTATCTGAGCGT----C------------------------------TGGGTGG-AACC--TCTGTGCTGCTAATG----------TT-AG---CTGATCCCTCTCCTCCCTCCCACTCGGTCCCTGCCCCCGCTCCCTTGGTGGCTGTTGCACTTTCCCCCGGGGCTCTGGA------------GTGTGCACACTTTTGGCA---------TAATA-CAACAAAATTGATTTG----------------------------------------------CAGCATGCATCATTATCAACATCGTCGTT |
| droWil1 |
scaffold_181089:335812-335995 + |
TTTTTTGAAATGTTTGTAAATGTGTGT----G----TGTTT------------GTGTGTGAAGAAGAA-TGTGTGTATGTGCTGACTAG--TTCAATAATG----C---C--------------------------------------------CAGAGAATGTTACAAGTTTAATGGAAGTTTTCAA-------------------------------------------ATAGAAAAG--------CTAAAAGGAA--------TTTAATTTTAGACAT-------CAGTTTCGAAGCATCGGCATCACAATCATCCCCATT |
| droVir3 |
scaffold_12963:15197471-15197651 + |
TATATATATAGATATATATATATATATATGCTGGGT----------------------TATATGA--------------------CTTTTG----------GG-TG---CTGATAAGAGGCG----TTCAGCTCAGTGC---------------CAGAGCCCAGCGGCTAATTGACAGCCCCACGGG-----------------CCATGCGGTTA---------AATTAAAA-CGAAAAA--------TCATTTGCCA--------GCA-GCAACAGC-AG----CAGCAGC--AGCGGCAGCAGC-----------------A |
| droMoj3 |
scaffold_6496:7093711-7093898 - |
TAGCAACTCGTTTCTGTTGCTGTATGT----GTAGCTGTAGCTGTATCTGTAGCTGTATCTGTGGCTG-TATCTGTATCTGGCAGCTGG----CAGCAGTA----G---C--------------------------------------------TGGTAGCTGCTGCTGGCTC--------TGTTGCT------GCCAGAGTATCTGTATCTGTGGCGCGTCAT-----------------------------TGT------------------------T-------GTGC--GTTGCCGTTGCCATCGTAACCAACGCCGTC |
| droGri2 |
scaffold_15126:1496468-1496644 - |
TACATCCACATCGCTGCTTCGGCTTAT----GTTGTGGTAA------------GTCATTAGAT--GCAG-ATGA--TTGGCGTCATCATCG----------CCTGCT--TCCACA--------------------------------------TCATAGTCAAGGGTATTCTTTTCTGCCGTCTC--AATGAGCGCTAGCG--------CCGATGAGATATCAA-----------------------------CGA------------------------G-------GTGC------TCTTCATCATCATCATCATCGTCATC |