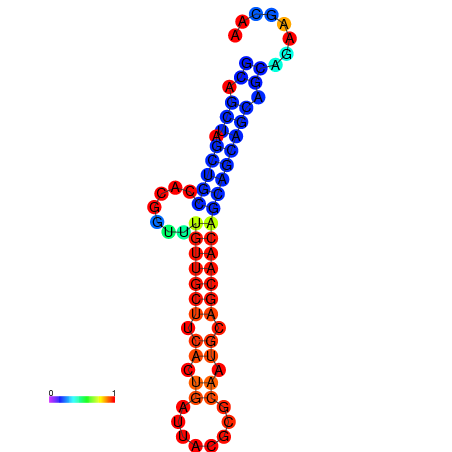
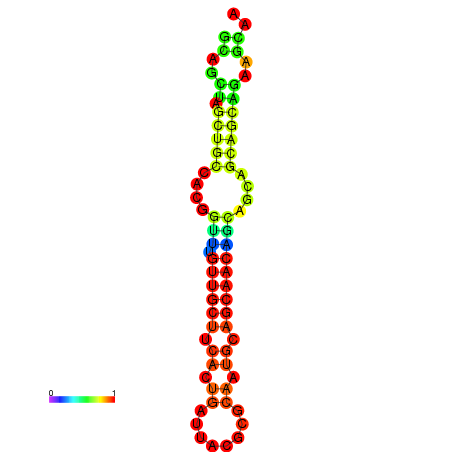
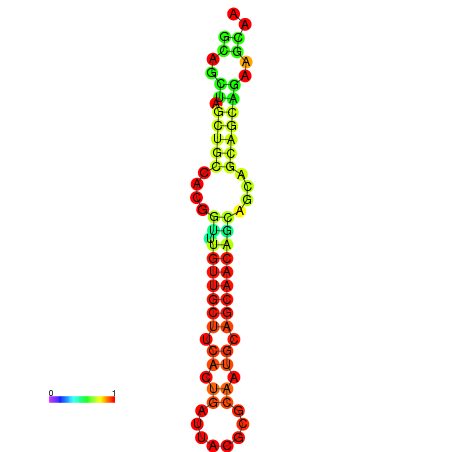
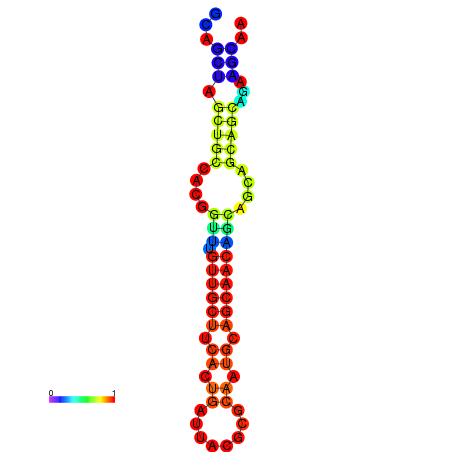
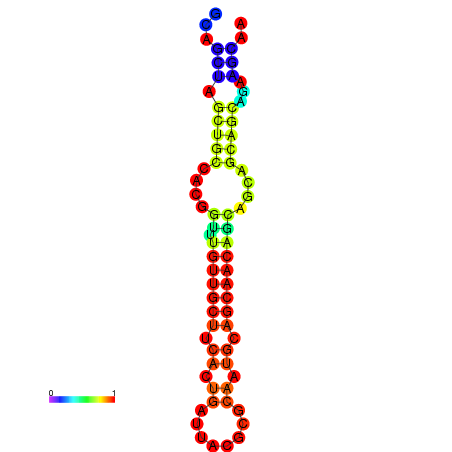
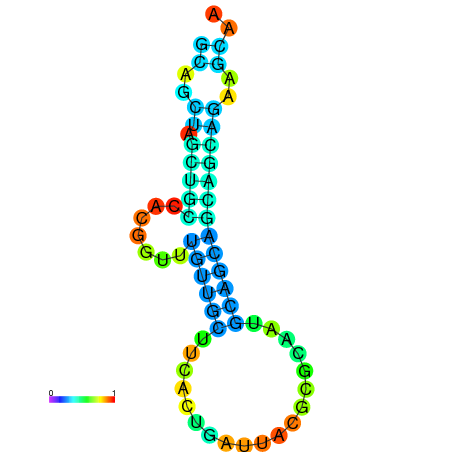
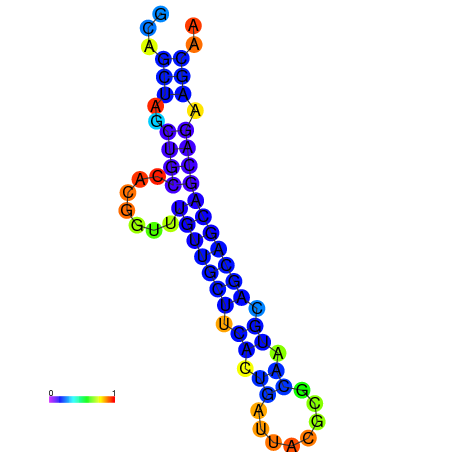
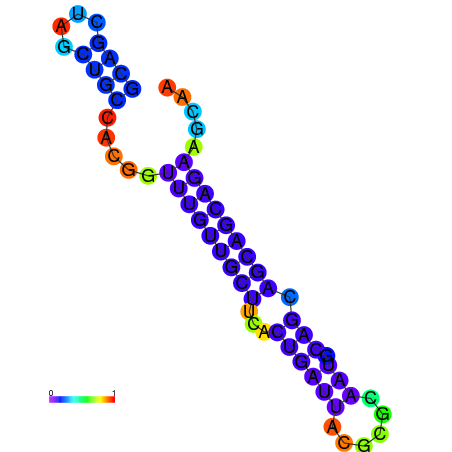
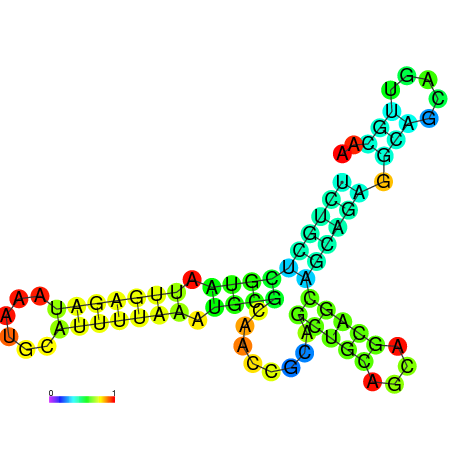
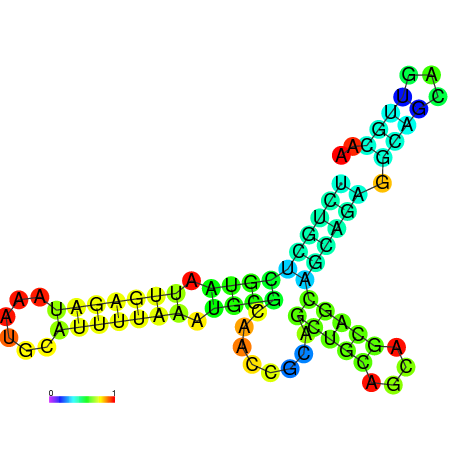
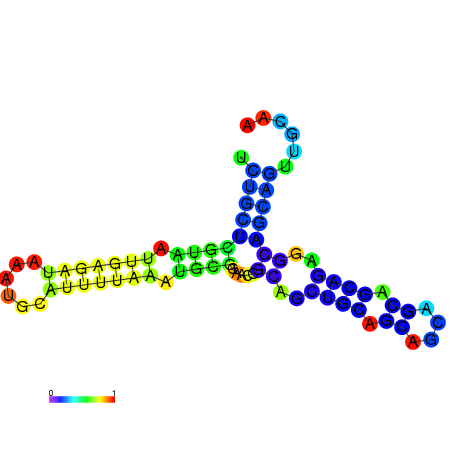
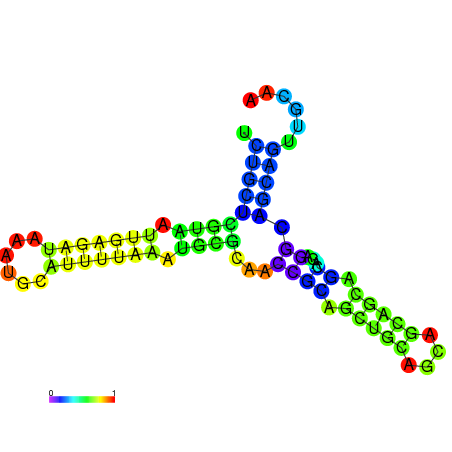
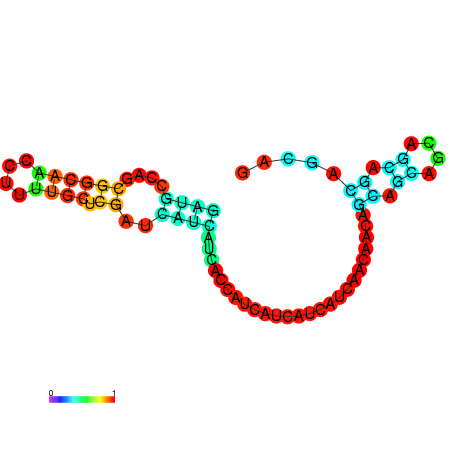
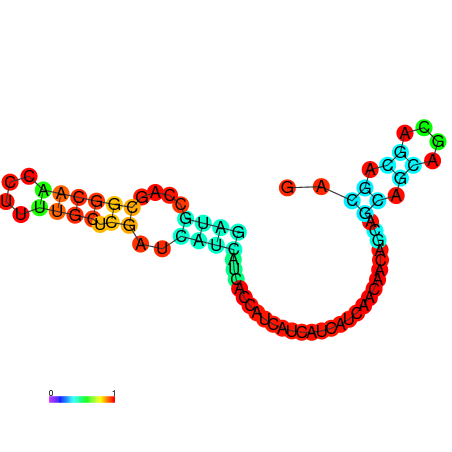
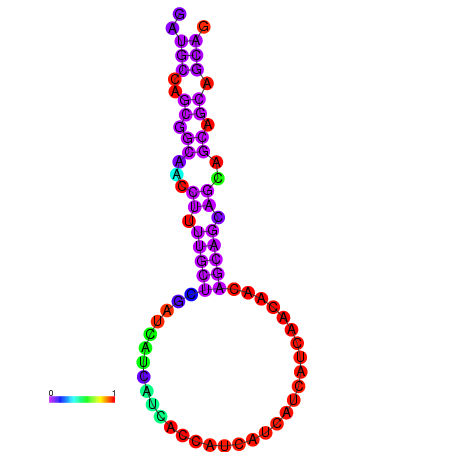
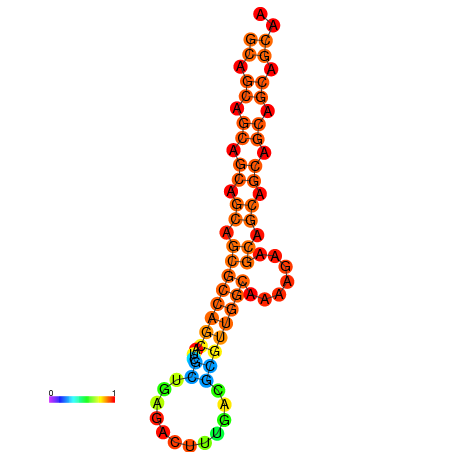
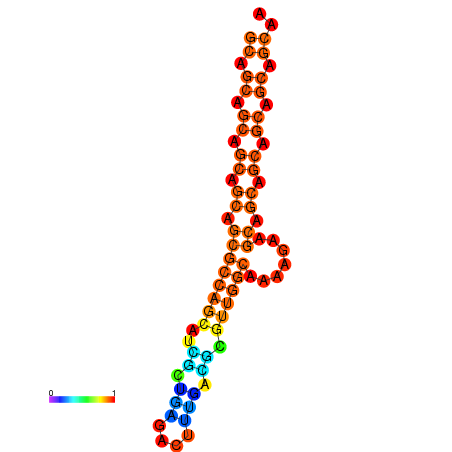
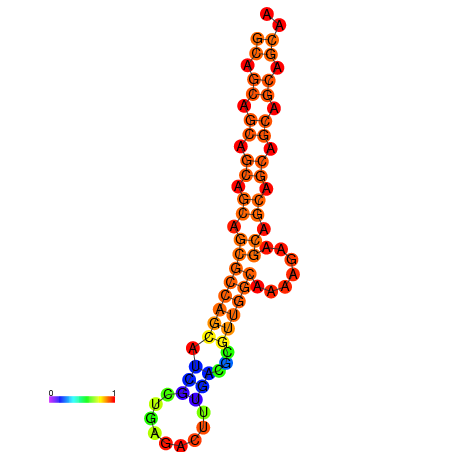
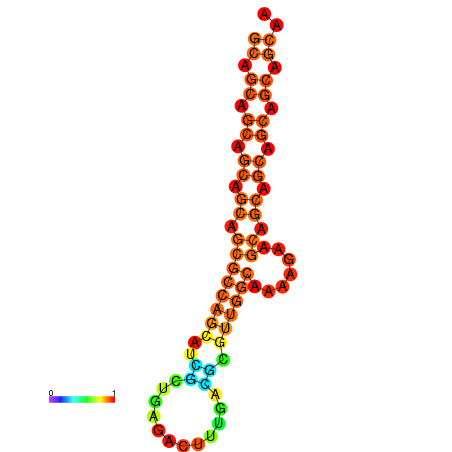
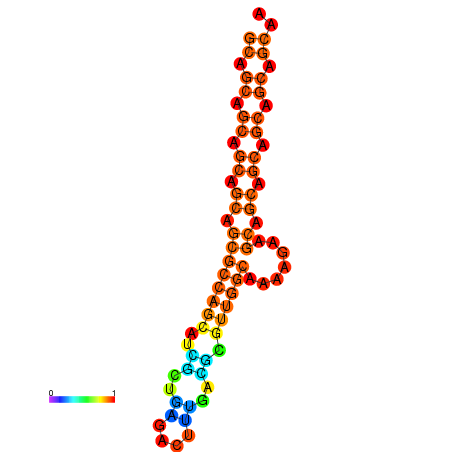
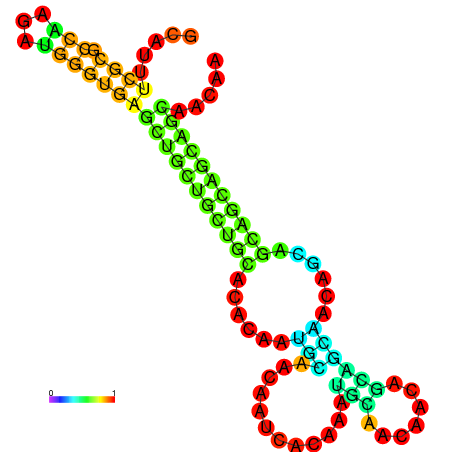
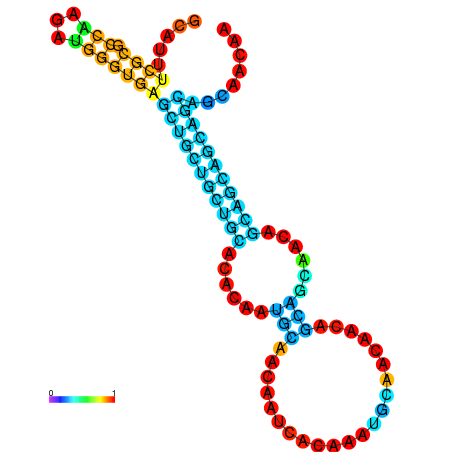
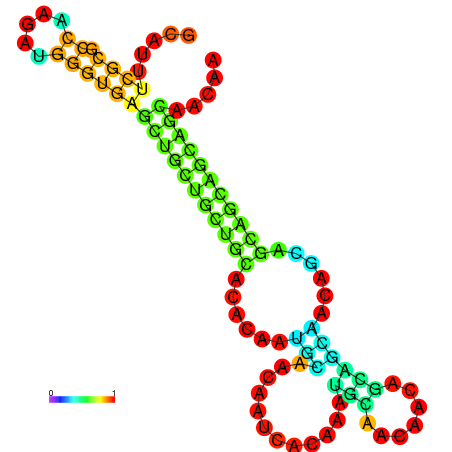
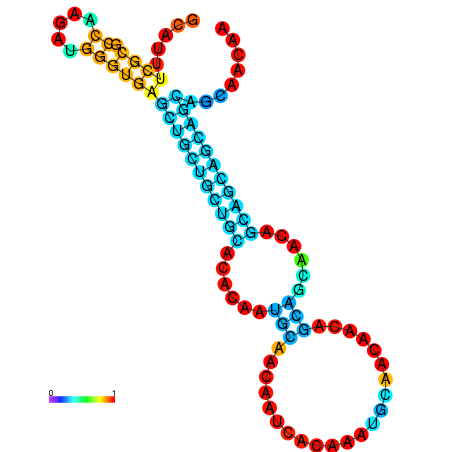
| dm3 |
chr3L:11545623-11545808 - |
AAACTGCATTCAAGT-----------TTTTCC-T-----------------------CAGAGC-----C-GCGACT----GCTGG---CGTGTTA-TTG------------------TTGC----TGTTGCAGCTAGCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAACAGCAGCA---------------GCAGC------A------------------------------------------------------------------GAAGCAACAGCAACT-AACTGCAGAGGAGCAATAGCAGCT-GAGAGTTCAGTTGCGAAAC-CGGCG |
| droSim1 |
chr3L:10929570-10929746 - |
AAACTGCATTCAAGT-----------TTTTCC-T-----------------------CAGAGC-----C-GCGACT----GCTGG---CGTGTTA-TTG------------------TTGC----TGTTGCAGCTAGCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGCAGCAG------------------------------------------------------------------------------------------------AAGCAACAGCAACT-AACTGCAGAGGAGCAATAGCAGCT-GAGAGTTCAGTTGCGAAAC-CGGCG |
| droSec1 |
super_0:3745423-3745605 - |
AAACTGCATTCGAGT-----------TTTTCC-T-----------------------CAGAGC-----C-GCGACT----GCTGG---CGTGTTA-TTG------------------TTGC----TGTTGCAGCTAGCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGCAGCAGCA---------------GCA---------------------------------------------------------------------------GAAGCAACAGCAACT-AACTGCAGAGGAGCAATAGCAGCT-GAGAGTTCAGTTGCGAAAC-CGGCG |
| droYak2 |
chr3L:11579958-11580167 - |
AAACTGCATTCAAGT-----------TTTTCC-T-----------------------CAGAGC-----C-GCGACT----GCTGG---CGTGTTA-TTG------------------TTGC----TGTTGCAGCTAGCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGCAGCAGAA---------------GCAGC---AGCAGCAGC----------------------------------AGTAGAAA------TAGCAGCA-----GTAGCAACAGCAACT-AACTGCAGAGGAGCAATAGCAGCT-GAGAGTTCAGTTGCGAAAC-CGGCG |
| droEre2 |
scaffold_4784:11545792-11545968 - |
A-ACTGCATTCAAGT-----------TTTTCC-T-----------------------CAGAGC-----C-GCGACT----GCTGG---CGTGTTA-TTG------------------TTGC----TGTTGCAGCTAGCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGCAGCAG------------------------------------------------------------------------------------------------AAGCAACAGCAACTGAACTGCAGAGGAGCAATAGCAGCT-GAGAGTTCAGTTGCGAAAC-CGGCG |
| droAna3 |
scaffold_13337:5830221-5830437 - |
AAACTGCATTCAAGTTTTGTTTTTGTTATTTTTT-----------------------CTTTGC--TTCT-GCAACTATT-GCTGGTTAGCTGCTAGTTG------------------CTGC----TGCTGCTGCT-GCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGTAGCAGTA---------------GCAGT---GGCA----------------------------------------------------GAG---GAGACAGAGGAGCCACTGCAAC--AACTGCAACAA---AACAGCAGCT-GAGAGTTCAGTTGCGAAAC-CGGCG |
| dp4 |
chrXR_group6:10126891-10127089 + |
AAACGGCATACAAGT-----------TTTTCC-TCGCCAAACACAAAGCGTTTTAGCCAGAGCTCTTCT-GCAACTATT-GCTGG-------TTG-TAG------------------TTGT----AGTT-------GCTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGCGGTGGCA---------------GCGGT---GGCA----------------------------------------------------GCG---GTGGCAGCGG-----TGGCA------------------GACGGCAGCCCCCGAGTTCAGTTGCAAAAC-CGGCG |
| droPer1 |
super_4:4953737-4953917 + |
TTTC-GTTTTC--GT-----------TTTCATTT--------------------------CGC-----T-GT--AA----ACAAG---C--GATT-TTG------------------TTGC----TCTTTCTGCTCGTAATTGAG-A-------TAAATGCATTTT---AAATGCGCAAC--CGCAGCTGCA------------------GCAGCAGCAGCA----------------------------------------------------GAGGCAGCA-----GTTGCAACAGCAGCT-AGCTTCAACT----GGTTGCAGTT-GCGTATGCAGATGCGATGCAAGGCG |
| droWil1 |
scaffold_180698:2057599-2057768 - |
ACACTGCAGACAAGTTTTAAATGCCTTTTTTT-T-----------------------CGTAGC-------------------TCT---CTTGTTG-TTGTTGTTGCTAA-GT--------C----TTCTGCAACTA-TTGCCACGGT-------TTGTTGCTTCACTGATTACGCGCAATGCAGCAGCAGCAGCG---------------GCAG---------------------------------------------------------------------------------CAGCA------------------AGCGGCAGCT-TCGAGTTCAGTTGCAAAAC-CGGCG |
| droVir3 |
scaffold_12855:885802-885977 - |
AGGATATGGGCGATT-----------GTCTTA-T-----------------------CAGTGG-----TGGTGCCG----GACCA---GCTGTTA-GTG------------------CCGT----TGCTGATGCCAGCGGCAACCTT-------TTGCTCGATCAT-CATCACCATCATC---------ATCAT--CAACAACAGCAGCAGCAGC------A----------------------------------------------------G--------------CAGCAGCAGCAACA-ACAACAACAGCAGCAACAGCAGCA-GCTACTGCAG--------------- |
| droMoj3 |
scaffold_6496:11355914-11356095 - |
AAACGAAATGAAACT-----------TTGTGC-G-----------------------CATAG--------GCAACCAACAGC-GG-------CGG-CAG------------------CAGC----AGCAGCAG------------------------------------------------------------------------------CAGC---AGCAGCAGCGCCAGCATCGCTGAGACTTTGACGCGTTGGCAAAAGAAGCAGCAGCAG------CA-----GCAGCAACAGCAAC--AACAGCC--GCAGCAGCAGCAGCA-GCGACGGCAGCAGCAGAAA-CGTCG |
| droGri2 |
scaffold_15110:4649694-4649883 + |
CAAT----------T-----------TTCTCA-T-----------------------CAGTTT-----C-T-------------------TGCCG-ATGATGAAGCAAATGTCATCGCTGCAGAATGTTGCATTTCGC-GCCAAGATGGGTGAGCTGCTGCTGC---------ACACAATGCAACAATCACAAATGCAACAACAGCAGCAACAGC------A----------------------------------------------------GCAGCAGCA-----GCAACAACAACA--------GC--AACAACAACAGGAGCC-G---ATGCAGCAGCA------ACCG |