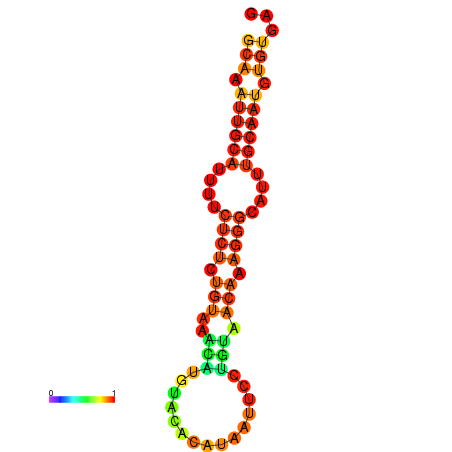
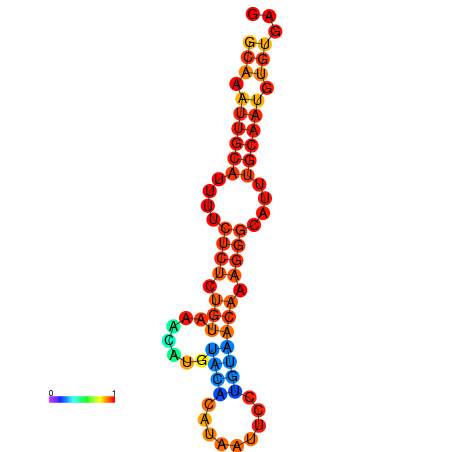
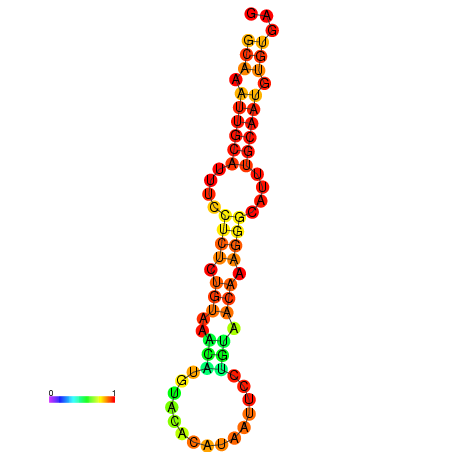
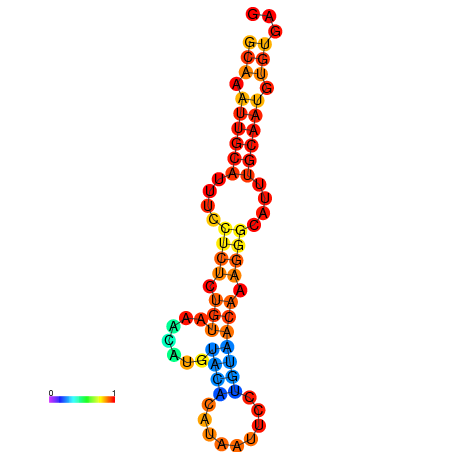
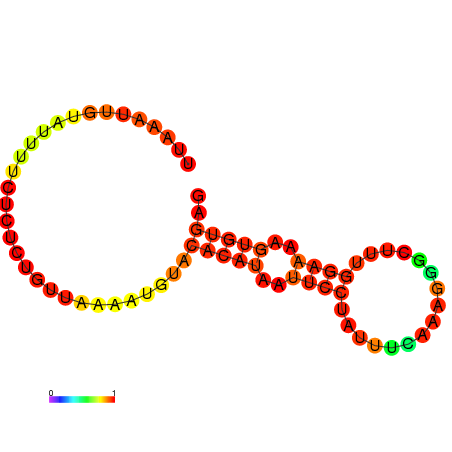
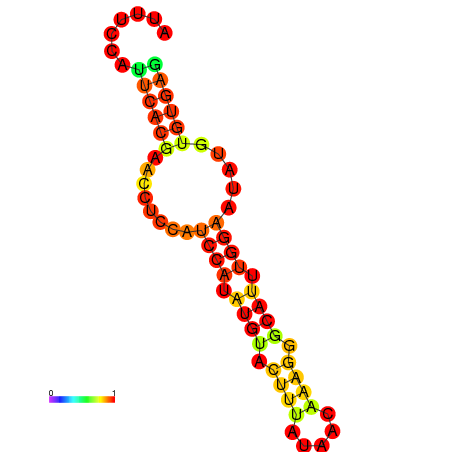
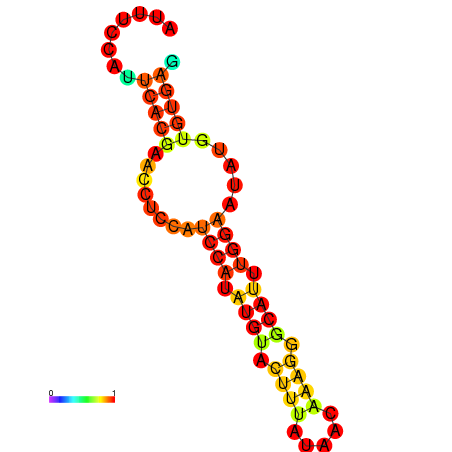
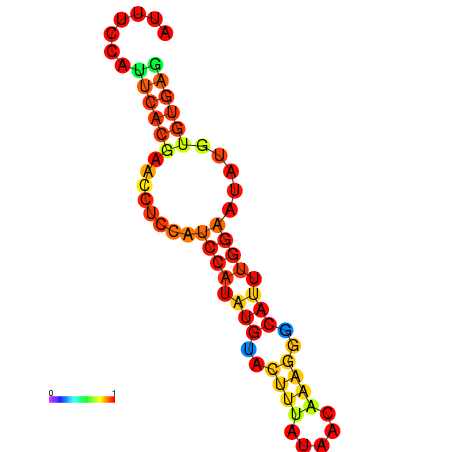
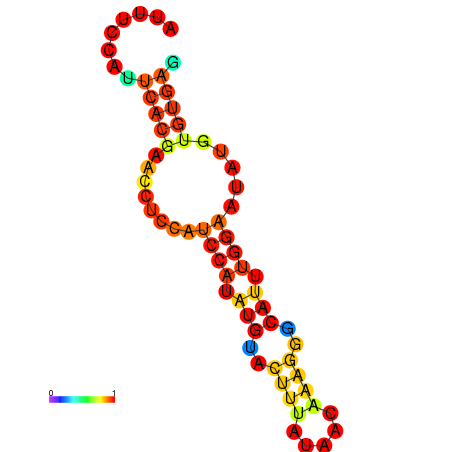
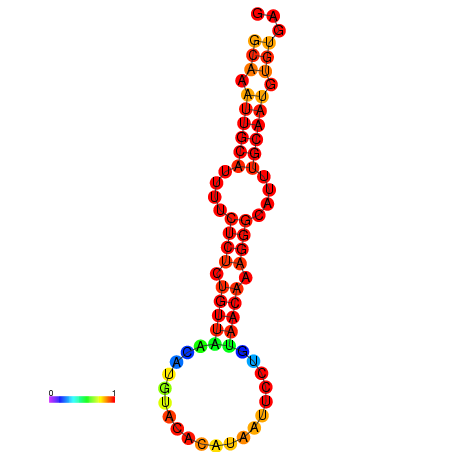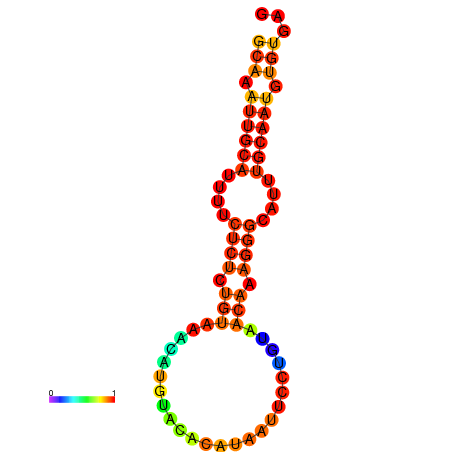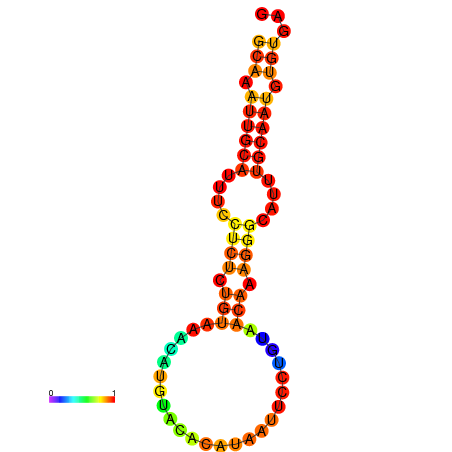| dm3 |
chr2R:5899582-5899719 - |
CATAGATATGGC--TAACTATAGCGTTGTCTCTTGCGCAAATTGCATTTTC---------TCTCTGTTAACATGTACACATAATTCCTGTAACAAAGGGCATTT-----GCAA--TGTGTGAGTCAGCAAA--CCGCATCCGCTTCCCGGC-CCCTCCT |
| droSim1 |
chr2R:4528976-4529111 - |
CGAGGAAATAGC--TAACTATAGCGTTCTCTCTTGCGCAAATTGCATTTTC---------TCTCTGTAAACATGTACACATAATTCCTGTAACAAAGGGCATTT-----GCAA--TGTGTGAGTCAGCAAA--CCGCATCCGCCCC-C--CGCCCTCCC |
| droSec1 |
super_1:3512309-3512447 - |
CGAGGAAATATCTTTAACTATAGCGTTCTCTCTTGCGCAAATTGCATTTCC---------TCTCTGTAAACATGTACACATAATTCCTGTAACAAAGGGCATTT-----GCAA--TGTGTGAGTCAGCAAA--CCGCATCCGCCCCTC--TGCCCTCCT |
| droYak2 |
chr2L:18527608-18527740 - |
CGGGAAACTCG-----ACTATAGCGTTCTCTCTTGTTTAAATTGTATTTTC---------TCTCTGTTAAAATGTACACATAATTCCTATTTCAAAGGGCTTTG-----GAAA--AGTGTGAGTCAGCAAA--CCGCATCCGCCCG-A--TTTCTTCCT |
| droEre2 |
scaffold_4929:8470755-8470887 + |
CGGTAAATTCG-----ACTAAAGCGTTCTCTCTTGCTTAAATTGTATTTTC---------TCTCTGTTAACATGTATACATATTTCCTGTAACAAAGGCCATTG-----GAAA--TGTGTGAGTCAGCAAA--CCGCATCCGCCCC-C--TTTCTTCCT |
| droAna3 |
scaffold_13266:17255662-17255760 + |
TGCTATA-TATCTGTACCTATA----------------------------------------------TGTATGTACGGGTG-TTTATGTAACAAAAGGCATTTTGGTGGAAA--TGCGTGAGTCAGCGGA--CCGCATTAT-------GC-CCTTTGT |
| dp4 |
chr3:6114287-6114365 - |
C-------------------------------------------------------------TCCATCCATATGTAC--------TTTATAACAAAGGGCATTT-----GGAATATGTGTGAGTCAGCACATGCCGCATCCGTG-----GC-CCCTTCC |
| droPer1 |
super_2:6317550-6317666 - |
TTTTGACA--GC-----TTTAAGCGTTCC----------------ATTTCCATTCACGAACCTCCATCCATATGTAC--------TTTATAACAAAGGGCATTT-----GGAATATGTGTGAGTCAGCACATGCCGCATCCGTG-----GC-CCCTTCC |
| droWil1 |
scaffold_180700:5327370-5327375 - |
CCC---------------------------------------------------------------------------------------------------------------------------------------------------------TTT |
| droVir3 |
scaffold_12875:417241-417243 - |
CCT------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droMoj3 |
scaffold_6496:5073155-5073192 + |
GAGACACTTA-------------------------------------------------------------------------------------ACTCCGTTC-----ACCT--TGTGCGA----------------------------C-TCCCTAA |
| droGri2 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------------- |