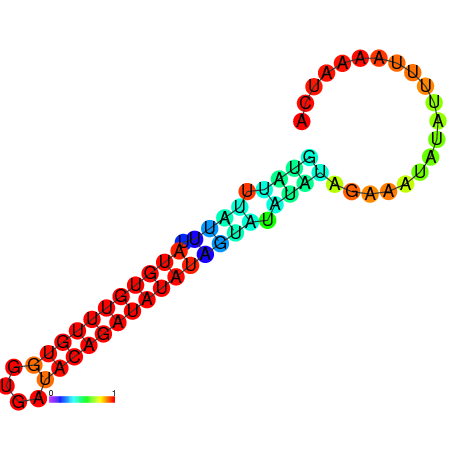
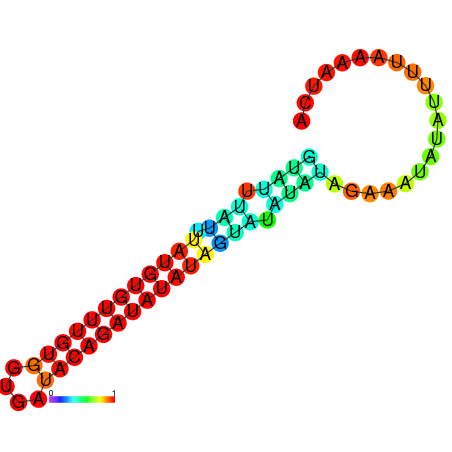
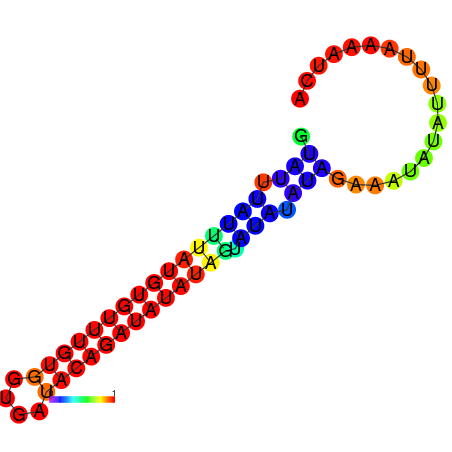
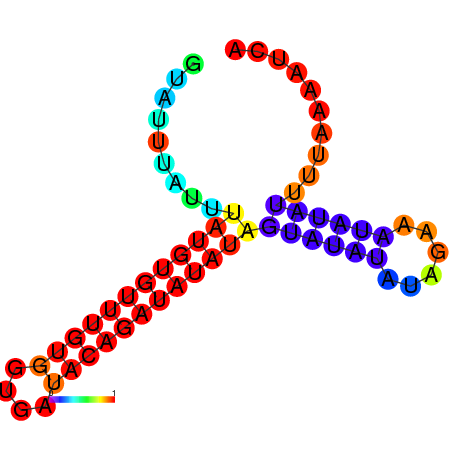
| dm3 |
chrX:1267640-1267803 + |
AGATATTACC--CAGAAGCGA-----------------------TATC------------------------CAATAGTAGCCAA-CTCTCTCG--CTCTC--T----------A----TGT--GTATGTATGTATCTTG---------------CTATCC---------------ATATA--TGTATATCCATATCAGAGAGCCAGGATTGGTTGAGACCATACG------ATATAACCCGAAACCACACTGG-CC-AGGAATA---------------------------------------------GCAAATCCA |
| droSim1 |
wjj76b12.b1:303-467 + |
AGATATTACC--CAGAAGCGA-----------------------TATC------------------------CAATAGTAGCCAA-CTCTCTCG--CTCTC--T----------A----TGT--GAATGTATGTATCTTG---------------CTATCC---------------ATATA--TGTATATCCATATCAGAGAGCCAGGATTGGTTGAGACCATACG------ATATAACCCGAAACCATACTGG-CC-AGGAATA---------------------------------------------GCAAATCCA |
| droSec1 |
super_10:1091085-1091248 + |
AGATATTACC--CAGAAGCAA-----------------------TATC------------------------CAATAGTAGCCAA-CTCTCTCG--CTCTC--T----------A----TGT--GAATGTATGTATCTTG---------------CTATCC---------------ATTTA--TGTATATCCATATCAGAGAGCCAGGATTGGTTGAGACCATACG------ATATAACCCGAAACCATACTGG-CC-AGGAATA---------------------------------------------GCAAATCCA |
| droYak2 |
chrX:1175521-1175702 + |
AGATATAACC--GAGAAGCAA-----------------------TACC------------------------CAATAGTAGCCAA-CTCTCTCGCTCTCTC--TGCGTCTCTCTA----TGT--GTATGTATGTATTTCG---------------CTATCC---------------ATATA--TGTATATCCATGTCAGAGAGCCAGGATCGGTTGAGACCATACTATATCTATATAACCCGAAACCATACTGG-CC-AGGAATA---------------------------------------------GCAAATCCA |
| droEre2 |
scaffold_4644:1251466-1251631 + |
AGATATAACC--CAAAAGAAA-----------------------TACC------------------------GAATAGTAGCCAA-CTCTCTCG--CTCTCTCT----------A----TGT--GTATGTATGTATTTCG---------------TTATCC---------------ATATA--TGTATATCCATGTCAGAGAGCCAGGATCGGTTGAGACCATACT------ATATAACCCAAAACCATACTGG-CC-AGGAATA---------------------------------------------GCAAGTCCA |
| droAna3 |
scaffold_13117:1856374-1856503 - |
--------------------------------------------------------------------------------------CTCTCTCT--CTCTC--TGTGTCTCTCTAAATGCAT--GTATGTATGTGTTGTA---------------TTATCC---------------GTATATATGTATATCCATGTTAGAGAGCCAGGACGGGTTGAGATT----------------GCCCAAGA----ACTGG-CCTAGGAATA---------------------------------------------GCAAAT-CA |
| dp4 |
chr2:10126391-10126530 + |
GAATAAT--CCATA-AACAAA-----------------------TATC------TA----------------T-GTAGTCA-CAT-CTATCTCT--CTCTC--T----------A----TAT--ATCTCTCTCTCTCTTT---------------CTTTAT---------------TTGTA--CATGTATTTATTTCATGGGTTTAGTTTTGC---AAATGGTATA------AAACATCTCCAAAATACATT------------------------------------------------------------------- |
| droPer1 |
super_2:4840479-4840636 - |
TAATATATTG--GAGTAGCAA-----------------------TATC------------------------TATTTGTATTCAG-ATTTGTAT--TTATT--T----------A----TGT--GTTTGTGGTGATACAG---------------ATATATA--------------GTATA--TATAGAAATATATTTTAAAATCA---TAGGTACATACAGTATA------TTGTATCCCTATCCAATGCATA-T-----GTGA---------------------------------------------ACGTACTTA |
| droWil1 |
scaffold_180700:3267577-3267741 + |
AAATATCATA--CCCA-AAAA-----------------------TATA------------------------TACTACTACACAACCTCTCTCT--CTCTC--T----------C----TAT--CTATCTATCTATCTTTGAACATAAAATGTACCTAT--ACTAAAACAAAAACTCTATA--TCTAAATCGTTGTGCT-TACCATTCATTTTCTACTACTATACT------ATATAGGGCGAAAC------------------------------------------------------------------------- |
| droVir3 |
scaffold_12855:2148540-2148695 + |
ATA----ACC--CAAAAGATA-----------------------CCTC------------------------AATTTGCATATAT-ATCTATGT--CTATA--T----------A----GAT--ATATACATACATATAT---------------ATATAC---------------ATATA--TATATATAAATATATGTGTGCGTGCATTA----TCGTCTTGTG------ACTAGGTCCAGAACTTTACAGG-TG-CGCAGTG---------------------------------------------TCGAAATCA |
| droMoj3 |
scaffold_6496:26031347-26031515 + |
TGATGCCACC--CAGC------------------------------CC------------------------CCACCGCCTCCTG-CTCCCTCC--CTCTC--TATGTTTATATG----TGT--ATATGTGTGT---------------------GTGTCT---------------GTGTG--TTTGTGTCCCTTC--------------------GATTGGCG------------------------TGCCTGAAT-GGCTGTGAAAACACAGCCAGTTGCAGCTGTATCCACAATCATTATCACAAAAGCAAACTAC |
| droGri2 |
scaffold_14853:6570585-6570746 + |
AAAAACAAAA--AACAAACGAAGTAACAGCCCAAGCCGACAATGGCTGAATATATATGTTATGTATTTGTATT-ATTGTC------------GT--GTATA--T----------A----TATCTATATATGTATATTACA---------------CTAT---------------------------------------------------------ATACTATATG-----TATATATATATAGACAATGTCTG-AT-GGCTATG---------------------------------------------CCAAAGCCA |