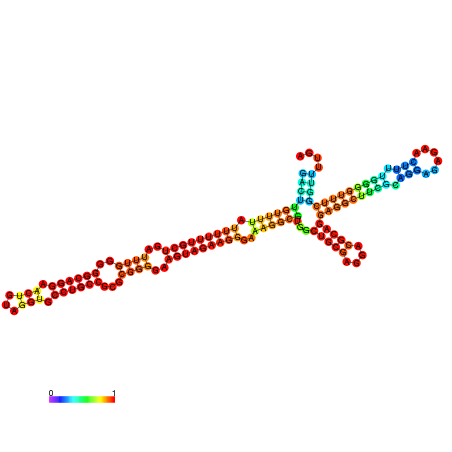
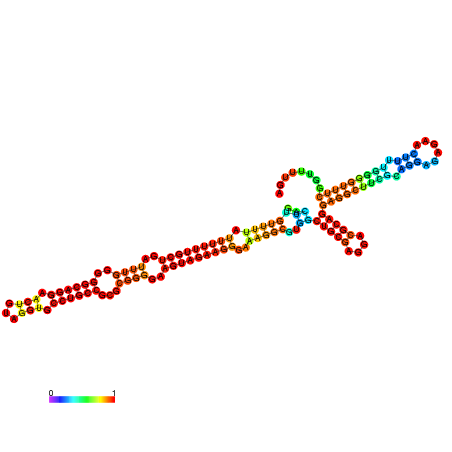
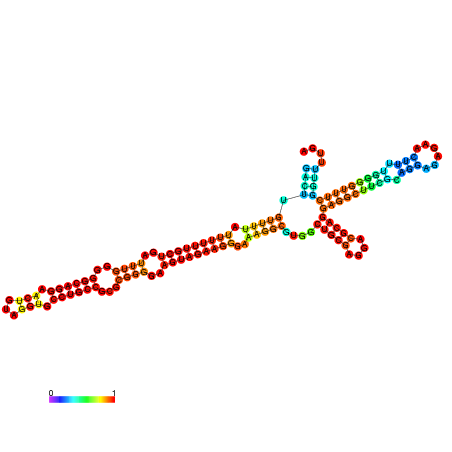
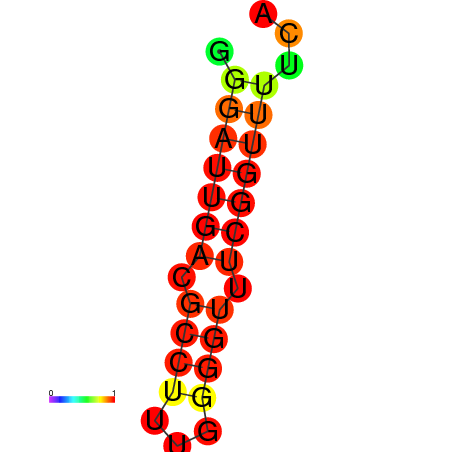
| dm3 |
chrX:463920-464051 + |
TGGAGTGACCAGCACAACACAAGGCCTGCCGCCCAAGAA----------------------------------------------------------------------------------------------------------------------ACGCAAGCCCAGAAGTCCGGCAGTC----------------GAGAA----------------------TCACGAGGGACTGG-G------ACCTTGGGCTTACGGTTCTCAGTTTGGCTTGACATGCATCAGCATCAT |
| droSim1 |
chrX_random:776904-777034 + |
CGGAGTGACCAGCACATCACAAGGCCTGGTGCCCAAGAA----------------------------------------------------------------------------------------------------------------------ACGCAAGCCCAGGAGTCCGGCAGTC----------------GAGAA----------------------TCACGAGGGACTGGGG------CCT-T-GGCTTACGGTTTTCATTTTGGCTTGACATGCATCAGCATCAT |
| droSec1 |
super_10:310837-310968 + |
CGGAGTGACCAGCACATCACAAGGCCTGGTGCCCAAGAA----------------------------------------------------------------------------------------------------------------------ACGCAAGCCCAGGAGTCCGGCAGTC----------------GAGAA----------------------TCACGAGGGACTGGG-------GCCTTGGGCTTACGGTTTTCATTTTGGCTTGACATGCATCAGCATCAT |
| droYak2 |
chrX:383437-383616 + |
TGCAATGCCCACCACAACACAAGGCCTGATGCCCGAGAAT-CGGTTTATTTCCTCCTCT------------------T-------------------------------------------------------------------------------CCGCAGGCCCAGGAGTCCGGCAGTC----------------GAGCAGTCGAGCTGTTCACCCGGTCAGTCCTT-GGTCCTTGGTCCTTGGGCCTTGGGCTTTCGGTTTTCAGTTTGGCTTGACATGCATCAGCATCAT |
| droEre2 |
scaffold_4644:409107-409303 + |
TGCAATGACCCTCACCACACAAGGCCCGATGCCCGGGAAT-CGGTTTATTTCCTCTTCT------------------T-------------------------------------------------------------------------------CCGCAGGCCCAGGAGTCCGGCAGTCCGACAGTCCAGCAGTCCGGCAGTCGAGTTGTCGAGTAGTCCAGTCATGAGGGGCTGGGGCCTTGGGCCTTGGGCTTACGGTTTTCAGTTTGGCTTGACATGCATCAGCATCAT |
| droAna3 |
scaffold_12929:357992-358198 + |
TGGAATGACC---ACAGCAGAAGGCCTTCTGCCAA-GACTTGGGTGTAGATATGAGTGTGTGTGGGTGTGGGTGTGTTTT--GGCCAGAATATGGTGGGTGATTGTGCAGGGAGGCGTGGCAGATGGGGG-GGCG-------GAGGATGAAGAAGAAGCAGTAGCAGAGGAG--------------------------------------------------------------------G------CCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| dp4 |
chrXL_group1e:196231-196421 + |
TGGAATGACC---ACGGCACAAGGCCT--TG-CCAAGACT-TGTTTTATTTTTTGCTGA------------------TTTGGGGGCAGGAACT-GTAGGTGCCT--GCCGCGCGGGGAAGTAGAAGGGAAAGGCGTGGCTGCGAG---GACGCAGGAGGCTT-------------CGCAGGA----------------GAG--------------------------------------AA------CTTTTGGGGTTTCGGTTTTGAGTTTGACTTGACATGCATCAGCATCAT |
| droPer1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
scaffold_180777:2769923-2769983 + |
CAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGATTGACG------CCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droVir3 |
scaffold_12970:10710181-10710231 + |
G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG------CCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droMoj3 |
scaffold_6473:10704148-10704209 - |
GGGG--------------------------------GGCTTGGG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG------CCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |
| droGri2 |
scaffold_14853:6069294-6069344 - |
G----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TG------CCTTTGGGGTTTCGGTTTTCAGTTTGACTTGACATGCATCAGCATCAT |