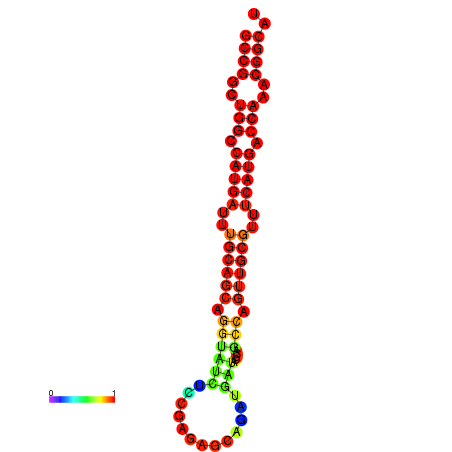
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19063148-19063296 - | TGCTCATGGCCCTGATGTCCGGTTTGAACCTTCTGTGCCGGCTGGCCATGATTTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGTTGCGTTTCATGACCAAACGGCATGTGAAGCGTGCCCTGCGGGACCTGACCATCGGC |
| droSim1 | chrX:14724057-14724205 - | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTACCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droSec1 | super_8:1352288-1352436 - | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droYak2 | chrX:17654811-17654959 - | TACTGATGGCCCTGATGTCTGGCCTAAATCTCCTCTGCCGGCTGGCCATGATCTGCAGCGGGTATCTTCGGGAGCAGATGATACGCAGCCAGTTGCGGTTCATGACCAAGCGGCATGTGAAGCGCGCCCTGCGCGACCTCACCATCGGC |
| droEre2 | scaffold_4690:9326382-9326530 - | TGCTGATGGCCCTGATTTCCGGCCTGAACCTCCTCTGCCGGCTGGCCATGATCTGCAGCAGGTATCTACGGGAGCAGATGATCCGCAGCCAGCTGCGGTTCATGTCCAAGCGGCATGTGAAGCGCGCCCTACGGGACCTCACCATCGGC |
| droAna3 | scaffold_12903:667658-667806 + | TCCTGATGGCCCTGATGTCCGGCCTGAATTTGGTCTGTCGCGTGGCGATGATTATCAGCAAGACCCTAAGAGAGCAGATGATCCGGAGCCAGTTGAGGTTCATGAAGAAGCGGCATGTCCAGAGGGCGCTGAGGGATCTGACGATCGGG |
| dp4 | chrXL_group1e:10756223-10756371 - | TCCTGATAGCCATCATATCCGGCGTGAATATCCTCTTCCGGCTGGCCATGCTGTCCATCAAGAGTCTGCGCGAGAAGATGATCCGCAGCCAGCTGTCCTTCATGACCAAACGACATGTGCAGCGGGCCCTGCGCGACCTCACCATCGGG |
| droPer1 | super_14:190427-190575 - | TCCTGATAGCCATCATATCCGGCGTGAATATCCTCTTCCGGCTGGCCATGCTGTCCATCAAGAGTCTGCGCGAGAAGATGATCCGCAGCCAGCTGTCCTTCATGACCAAACGACATGTGCAGCGGGCCCTTCGCGACCTCACCATCGGG |
| droWil1 | scaffold_181096:8247011-8247159 - | TGCTGATGGCCATCATGTCGGGCCTCAATCTACTCTGTCGCCTGGCCATGATGCTGAGCAAGAGTGTGCGTGAGCAAATGATTCGCAGTCAATTGCGTTTTATGCCCAAACGGCATATCCAACGAGCTCTCAGGGATCTAACCATTGGC |
| droVir3 | scaffold_12726:998444-998592 - | TGCTGATGGCCTTCATGTCCGGCCTGAACCTGCTCTGCCGTCTGGCCATGATGCTCAGCCGGACGCTGCGTGAGCTGATGATACGCAGCCAATTGCGCTTCATGACCAAGCAGCATGTACAGCGCGTTTTCCGGGATCTAACCATCGGT |
| droMoj3 | scaffold_6473:9166585-9166733 + | TGCTGATGGCCTTCATGTCCGGCCTGAATCTGCTCTGTCGCCTGGCCATGATGCTCAGCCGGACGCTGCGCGAGCTGATGATACGCAGCCAATTGCGCTATATGACCAAACAGCATGTGCAGCACGTCTTCAGGGATCTGACCATCGGC |
| droGri2 | scaffold_14853:5433396-5433544 - | TCCTCATGGCGATCATCGCTGGCCTAAATCTGCTCTGTCGCCTAACCATGATGGTCAGCCGGACGCTGCGCGAACTGATGATTCGCAGCCAGTTGCGATTCATGACCAAAGAGCATGTGCAGCGCATCTTTAGCCATTTAACCATCGGC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:19063148-19063296 - | TGCTCATGGCCCTGATGTCCGGTTTGAACCTTCTGTGCCGGCTGGCCATGATTTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGTTGCGTTTCATGACCAAACGGCATGTGAAGCGTGCCCTGCGGGACCTGACCATCGGC |
| droSim1 | chrX:14724057-14724205 - | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTACCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droSec1 | super_8:1352288-1352436 - | TGCTCATGGCCCTGATGTCCGGCTTGAACCTTCTGTGCCGGCTGGCCATGATCTGCAGCAGGTATCTCCGAGAGCAGATGATACGCAGCCAGCTGCGTTTCATGACCAAGCGGCATGTGAAGCGTGCCCTGCGTGACCTGACCATCGGC |
| droYak2 | chrX:17654811-17654959 - | TACTGATGGCCCTGATGTCTGGCCTAAATCTCCTCTGCCGGCTGGCCATGATCTGCAGCGGGTATCTTCGGGAGCAGATGATACGCAGCCAGTTGCGGTTCATGACCAAGCGGCATGTGAAGCGCGCCCTGCGCGACCTCACCATCGGC |
| droEre2 | scaffold_4690:9326382-9326530 - | TGCTGATGGCCCTGATTTCCGGCCTGAACCTCCTCTGCCGGCTGGCCATGATCTGCAGCAGGTATCTACGGGAGCAGATGATCCGCAGCCAGCTGCGGTTCATGTCCAAGCGGCATGTGAAGCGCGCCCTACGGGACCTCACCATCGGC |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
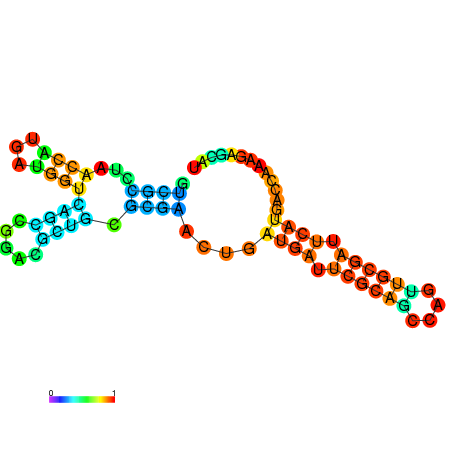
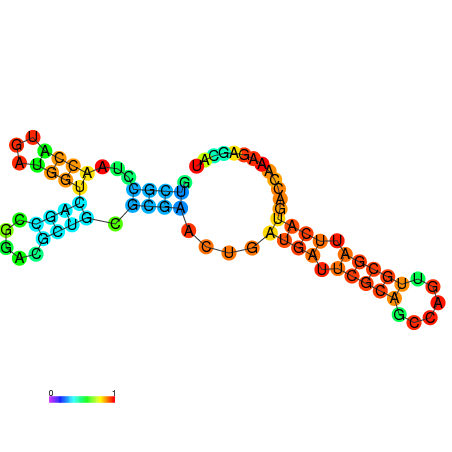
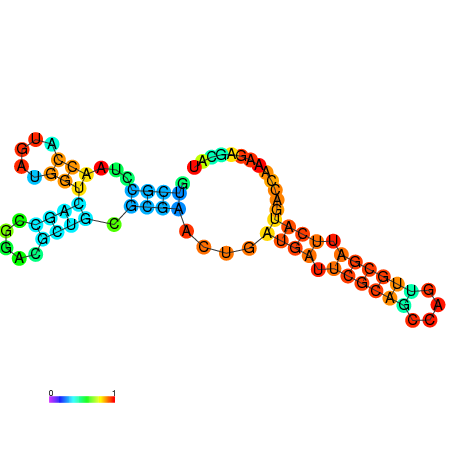
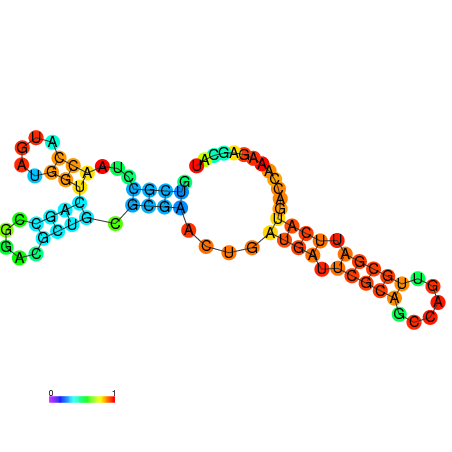
| dG=-30.4, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-29.7, p-value=0.009901 | dG=-29.6, p-value=0.009901 | dG=-29.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
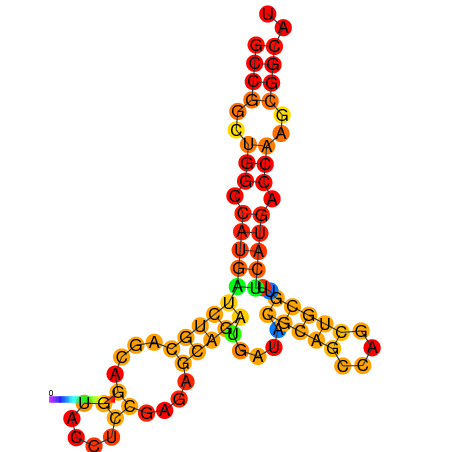
| dG=-32.1, p-value=0.009901 | dG=-31.8, p-value=0.009901 | dG=-31.1, p-value=0.009901 | dG=-30.9, p-value=0.009901 |
|---|---|---|---|
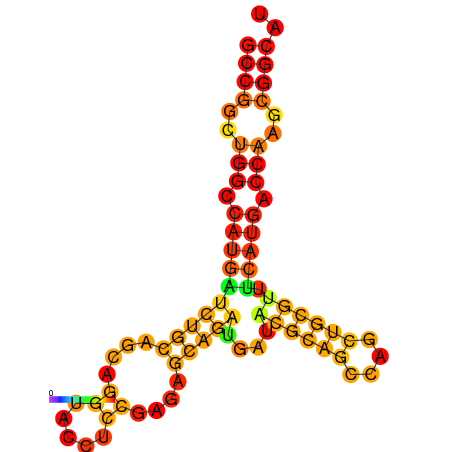 |
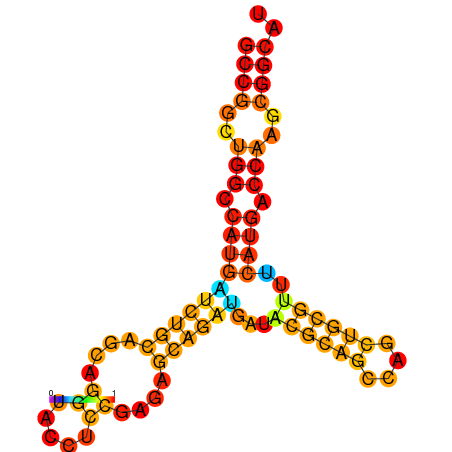 |
 |
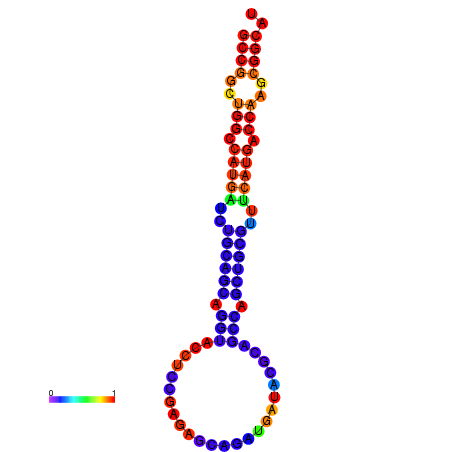 |
| dG=-32.5, p-value=0.009901 | dG=-32.3, p-value=0.009901 | dG=-32.1, p-value=0.009901 | dG=-31.8, p-value=0.009901 | dG=-31.8, p-value=0.009901 |
|---|---|---|---|---|
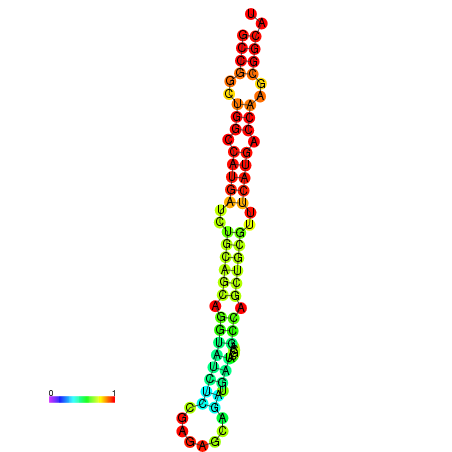 |
 |
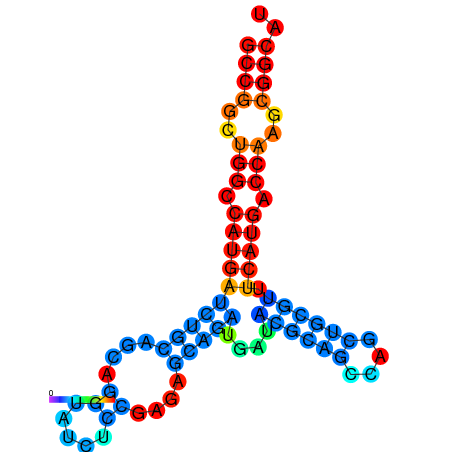 |
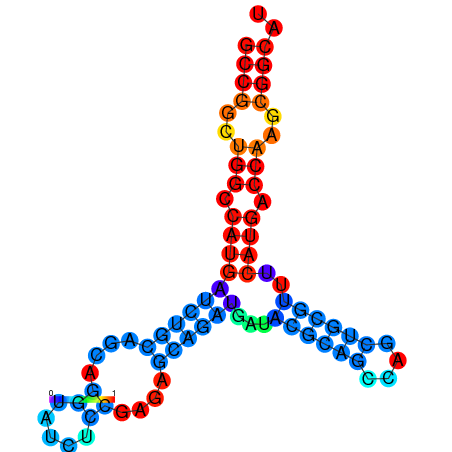 |
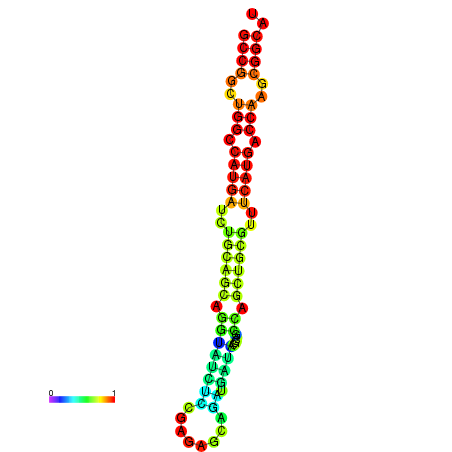 |
| dG=-37.0, p-value=0.009901 |
|---|
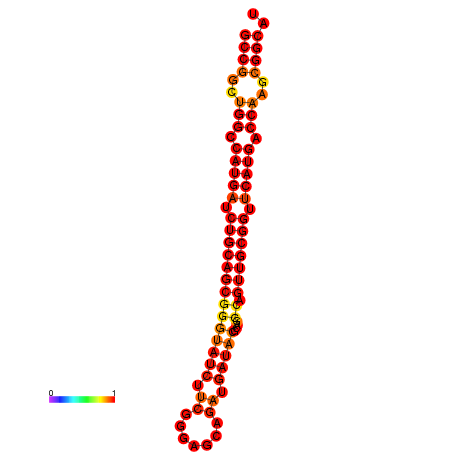 |
| dG=-37.9, p-value=0.009901 | dG=-37.1, p-value=0.009901 | dG=-37.1, p-value=0.009901 | dG=-36.9, p-value=0.009901 | dG=-36.9, p-value=0.009901 |
|---|---|---|---|---|
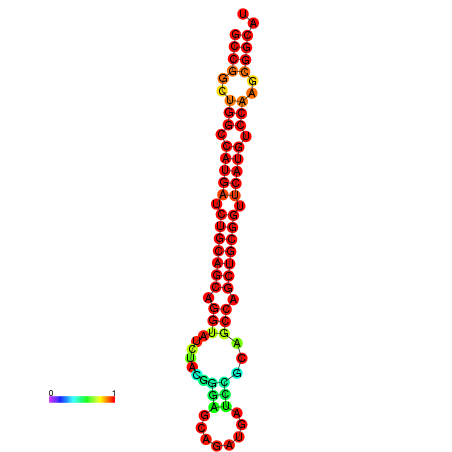 |
 |
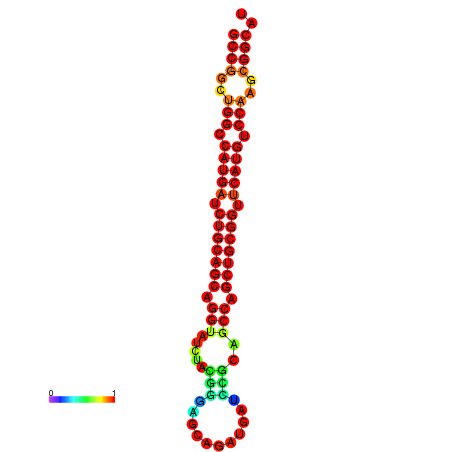 |
 |
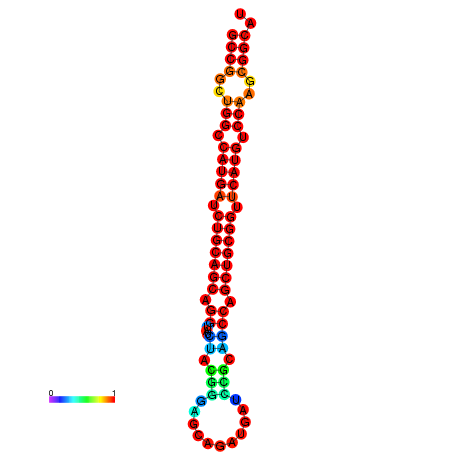 |
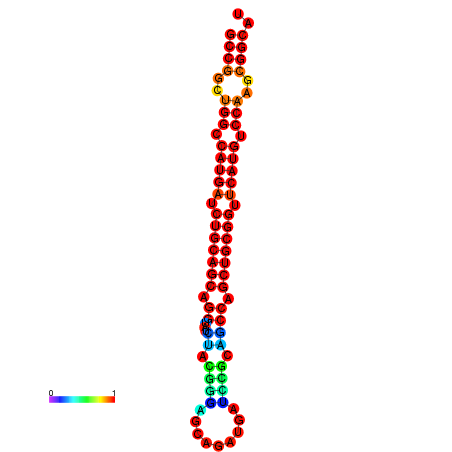
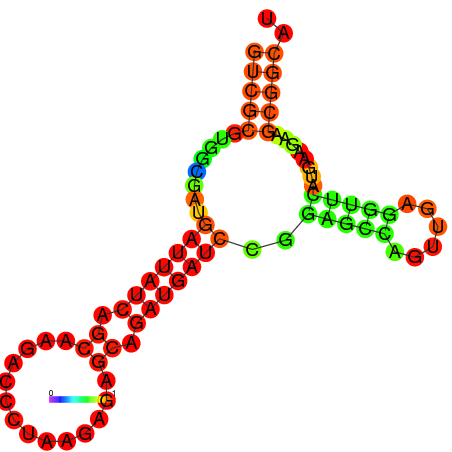
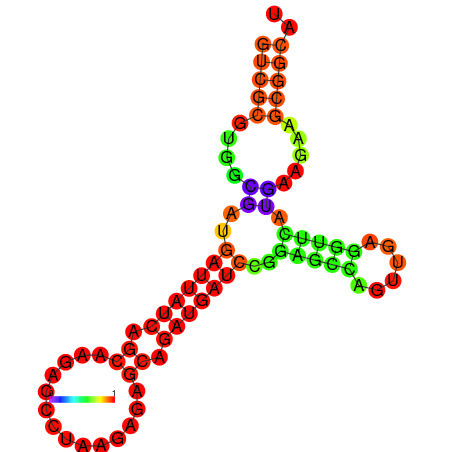
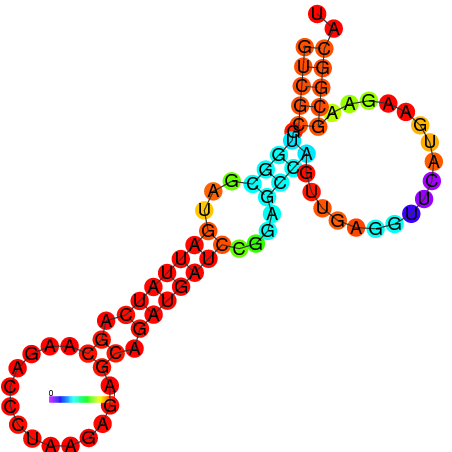
| dG=-19.7, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.5, p-value=0.009901 | dG=-18.9, p-value=0.009901 | dG=-17.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
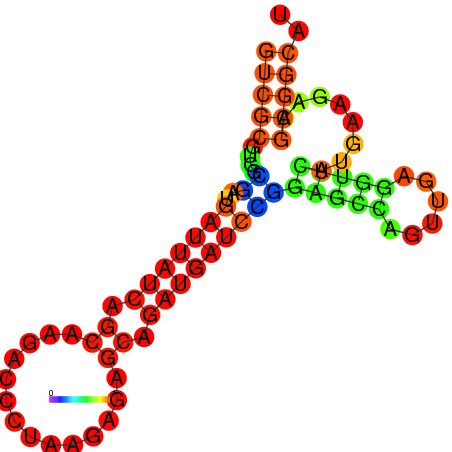
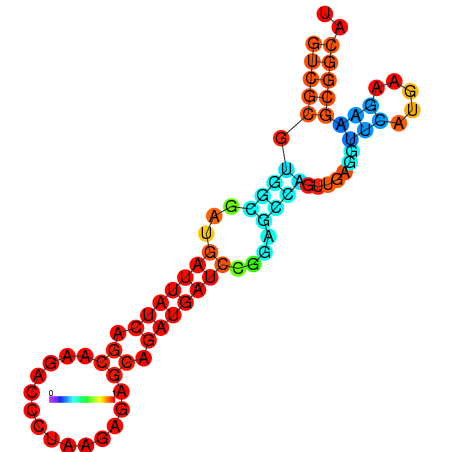
| dG=-19.3, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 |
|---|---|---|---|---|
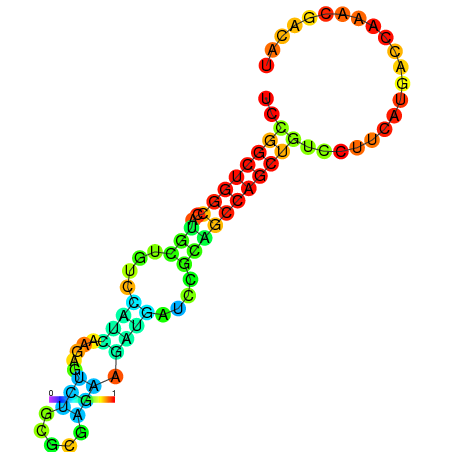 |
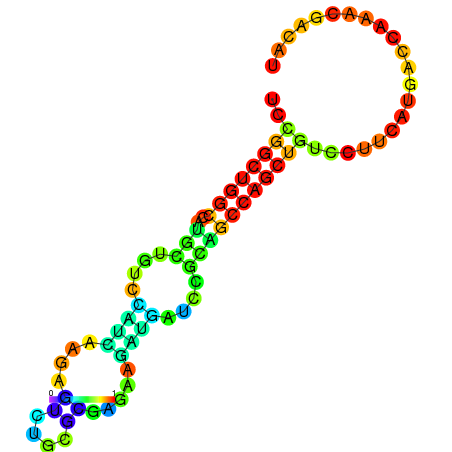 |
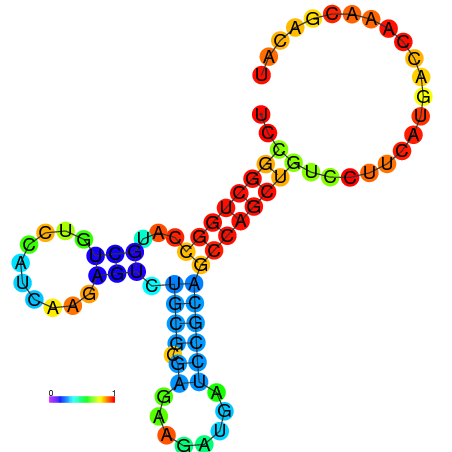 |
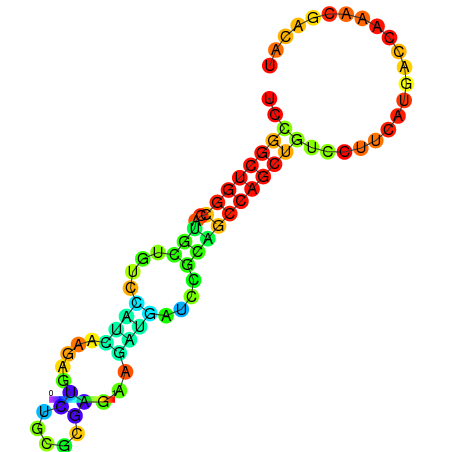 |
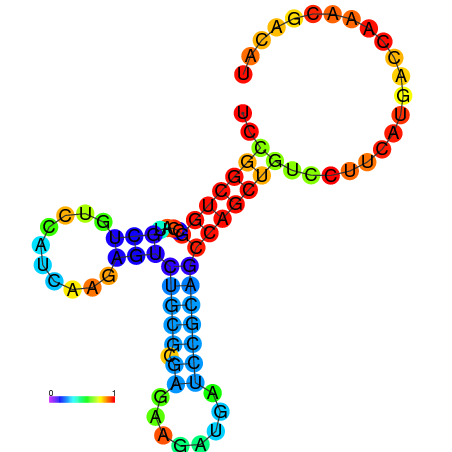 |
| dG=-19.3, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-19.1, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 |
|---|---|---|---|---|
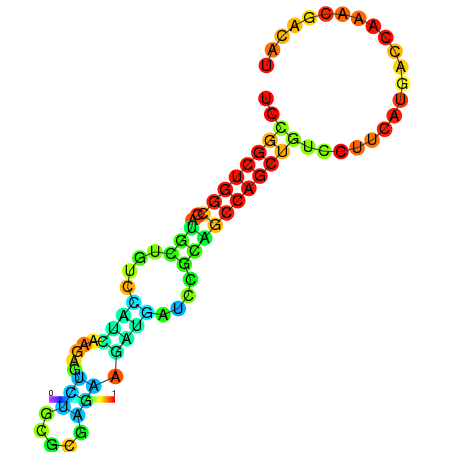 |
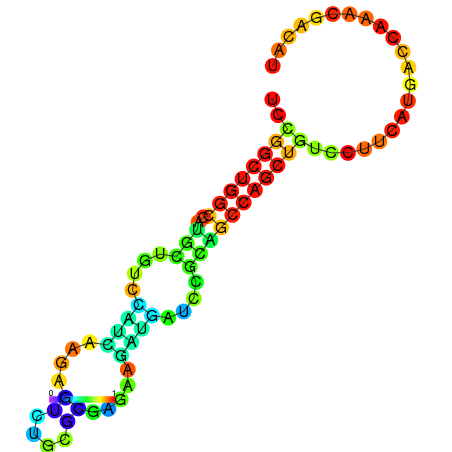 |
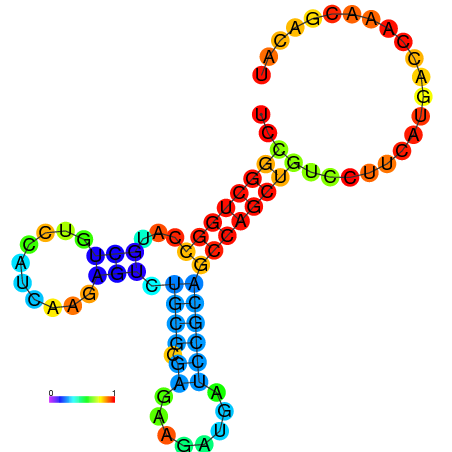 |
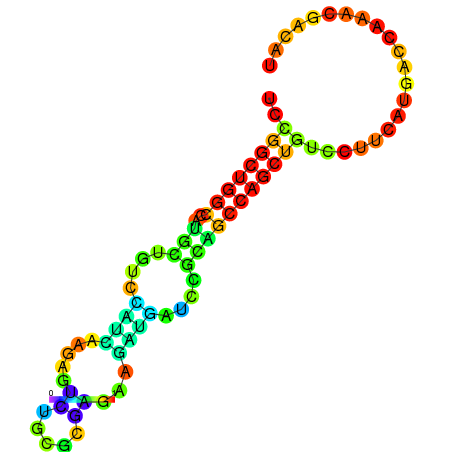 |
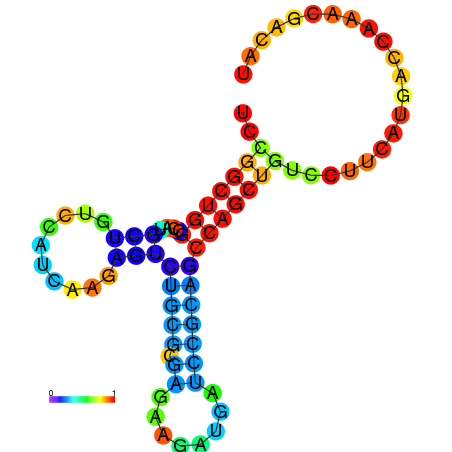 |
| dG=-21.8, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-21.1, p-value=0.009901 | dG=-21.0, p-value=0.009901 |
|---|---|---|---|---|
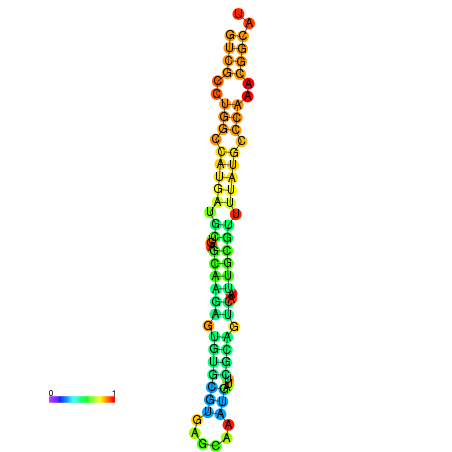 |
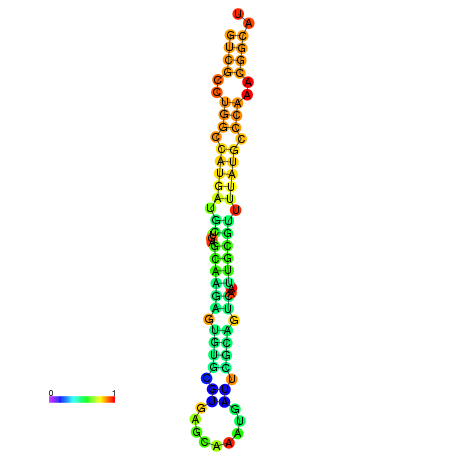 |
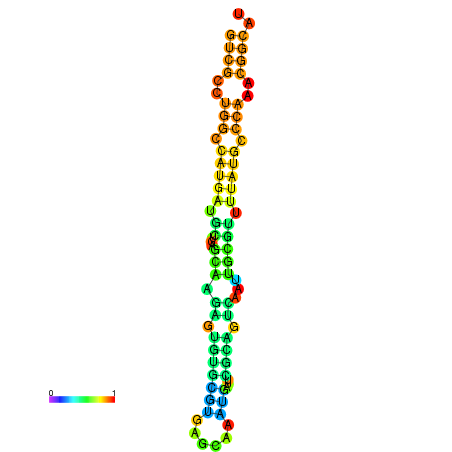 |
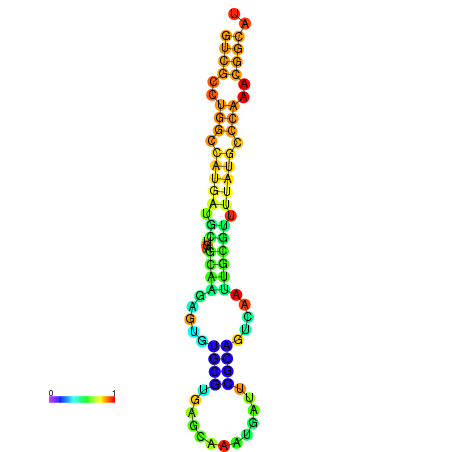 |
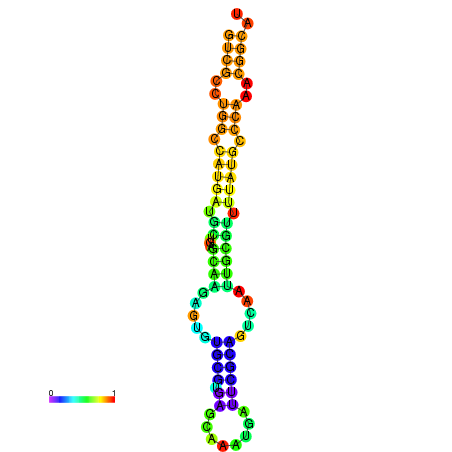 |
| dG=-28.8, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.3, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-27.6, p-value=0.009901 |
|---|---|---|---|---|---|
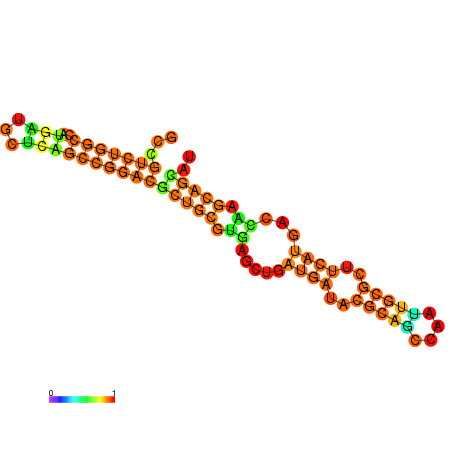 |
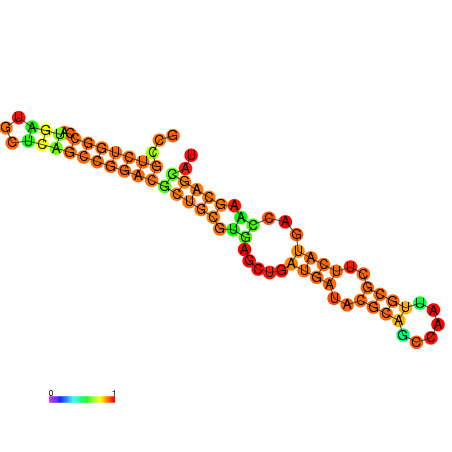 |
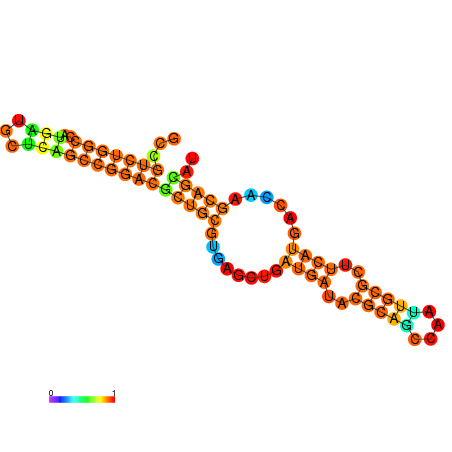 |
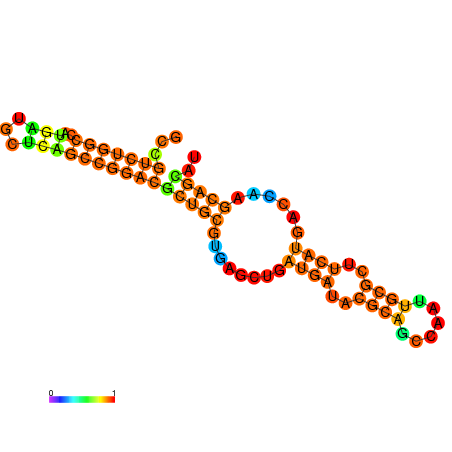 |
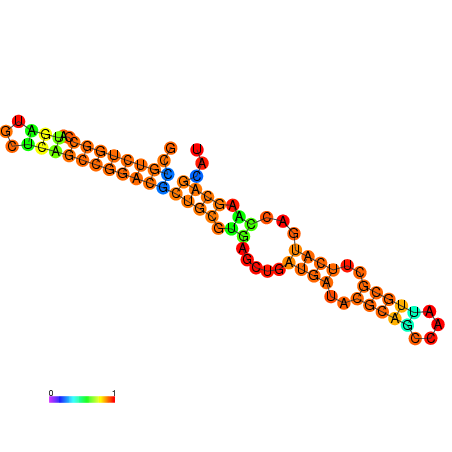 |
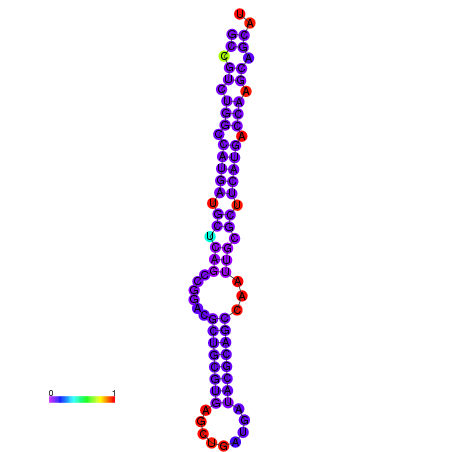 |
| dG=-21.1, p-value=0.009901 | dG=-20.4, p-value=0.009901 | dG=-20.2, p-value=0.009901 | dG=-18.6, p-value=0.009901 |
|---|---|---|---|
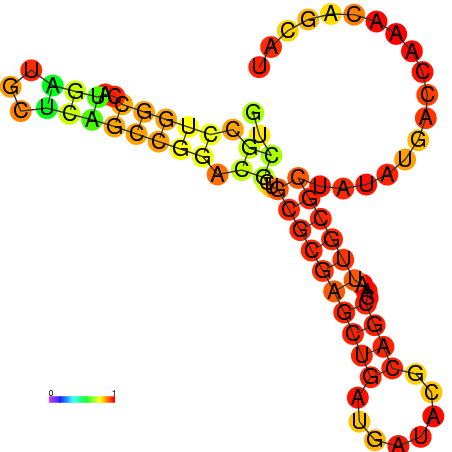 |
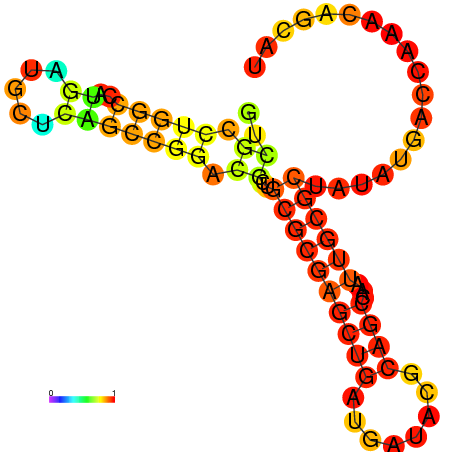 |
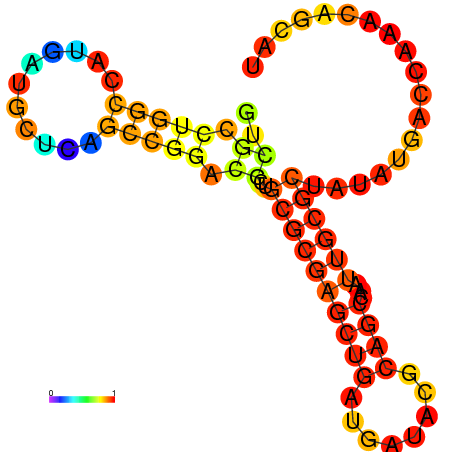 |
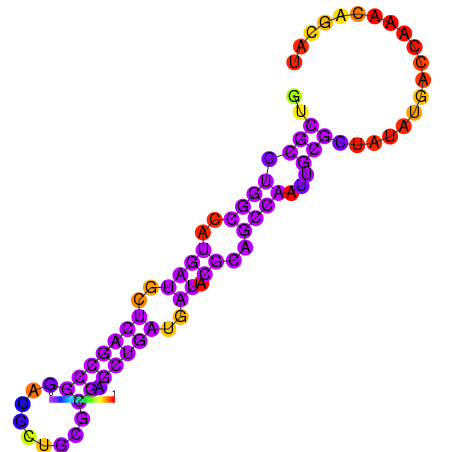 |
| dG=-19.6, p-value=0.009901 | dG=-19.6, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-18.8, p-value=0.009901 | dG=-17.4, p-value=0.009901 |
|---|---|---|---|---|---|
 |
 |
 |
 |
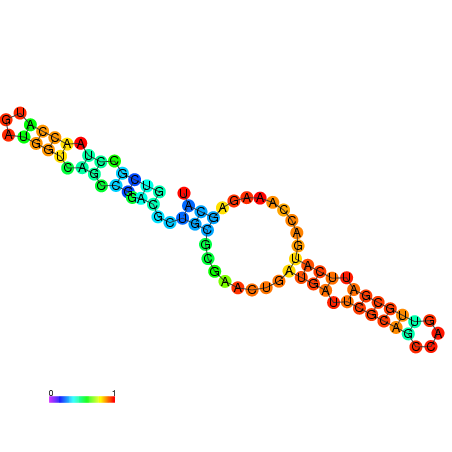 |
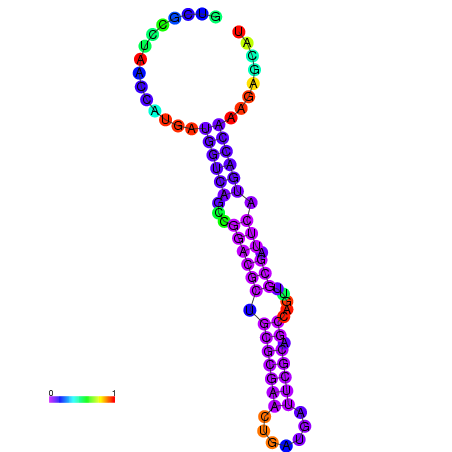 |
Generated: 03/07/2013 at 06:07 PM