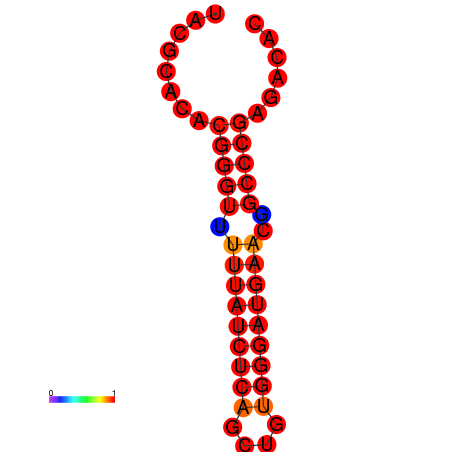
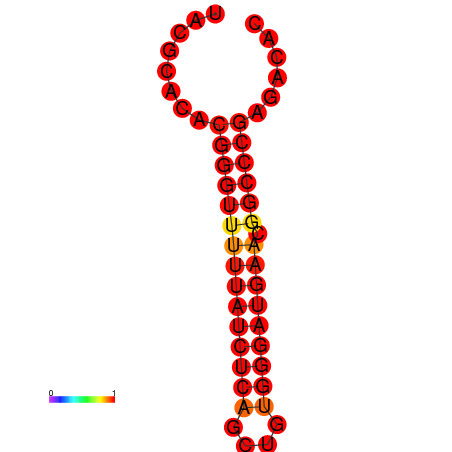
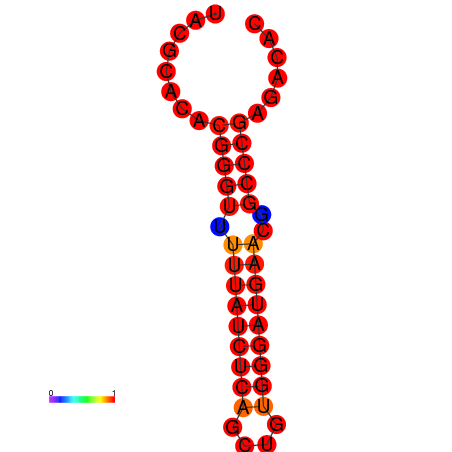
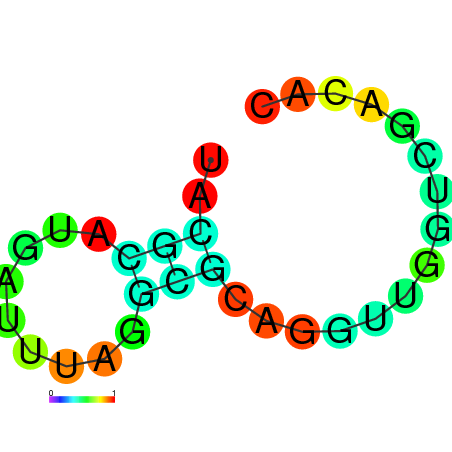
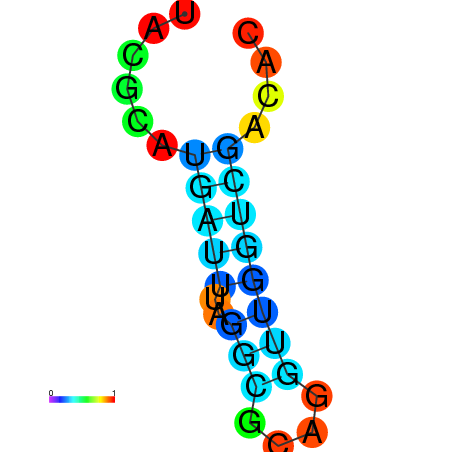
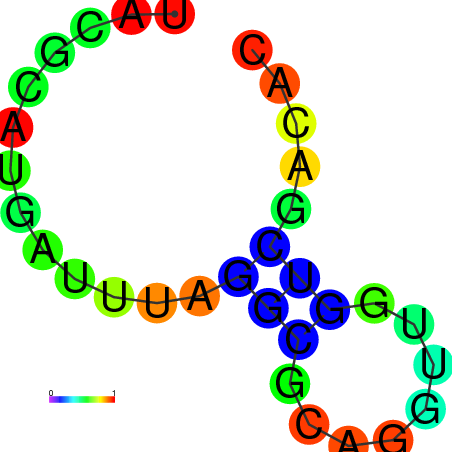
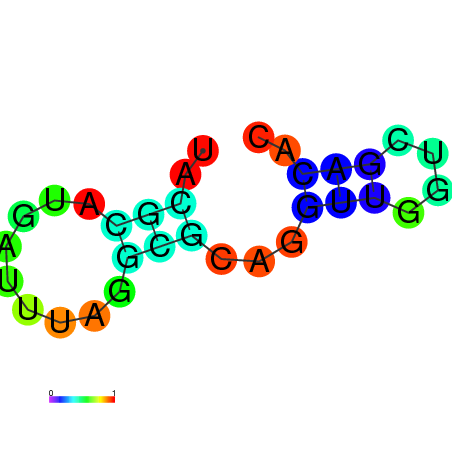
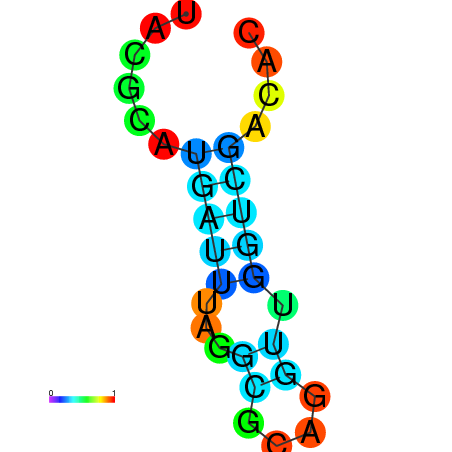
| dm3 |
chrX:15213971-15214085 - |
CTTTTTTTT--------------CTCTGCTC----GT-----------------------T--GTA--------TTTTACGCA--CTCTGCTGGCAGCG---CTGTCGGC---GTCGCTGCCAGCTGCCAGCAGCGAGCGGTACACTT--------GCAACTATC-----CTGT-GCACCCTACAATT |
| droSim1 |
chrX:11736931-11737042 - |
CTTTTTTTTTTTT----------CTCTGCTC----GT-----------------------T--GCA--------TTTTACGCA--CTCTGCTGGCAGCG---ACGCCGAC---AGCGCTGCCA-------GCAGCGAGCGGTACACTT--------GCAACTATC-----CTGC-GCACCCTACAATT |
| droSec1 |
super_56:101687-101796 - |
CTTTTTTTTT--T----------CTCTGCTC----GT-----------------------T--GCA--------TTTTACGCA--CTCTGCTGGCAGCG---ACGCCGAC---AGCGGTGCCA-------GCAGCGAGCGGTACACTT--------GCAACTATC-----CTGC-ACACCCTACAATT |
| droYak2 |
chrX:9414156-9414285 - |
TTTTTTT----------------CTCTGCTC----GTTTTTCTCTCTTTTTCTTCTCGCCT--GCA--------TTTTACGCA--CTCTGCTAGCAGCG---ACGCCGAC---TGCGCTGCCA-------ACAGCAAGCGGTACACTT--------GCAACTATT-----CTAT-GCACCCTACAATT |
| droEre2 |
scaffold_4690:11807313-11807440 + |
TTTTTT-----------------CTCCGCTC----GTTTTTCTCTCCTTTTCTGCTCGCCT--GCA--------TTTTACGCA--CTCTGCTGGCAGCG---ACGCCGAC---TGCGCTGCAA-------ACAGCGAGCGGTACACTT--------GCAACTATT-----CTGT-GCACCCTACAATT |
| droAna3 |
scaffold_13117:1427273-1427395 - |
ATTTTTCTTC---CCATGCAACC-TGTGTCC----AT-----------------------TGTACG--------TTTTACGCA--CTCTCC--CCAGCA---ACACCGCC---GCCGATAGCG-------ACTGCGACGAGTACGTTG--------GCAGCGGCACTTGACTCT-TTTTGCGGCAATT |
| dp4 |
chrXL_group1e:12152468-12152588 - |
CTTTAGCTTT---TTG--------------CGTTTGCGTTTGCCCCGATTTCAATGCA-AT--GTG--------TTGTACGCA--CACGGGTT---TTT---ATCTCAGC---TGTGGGATGA-------ACGGC--CCGAGACACTT--------GC--CGATG-----CGGC-GA-CCC--CAGGC |
| droPer1 |
super_14:1456573-1456693 + |
CTTTAGCTTT---TTG--------------CGTTTGCGTTTGCCCCGATTTCAATGCA-AT--GTG--------TTGTACGCA--CACGGGTT---TTT---ATCTCAGC---TGTGGGATGA-------ACGGC--CCGAGACACTT--------GC--CGATG-----CGGC-GA-CCC--CAGGC |
| droWil1 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12928:4095173-4095244 - |
CTCTTTATTG---CAA--------------------------------TGCGAC-----------------------TACGCATGATTTAGGCGCAGGT---TGGTCG----------------------------------ACACTT--------GCATTGGTT-----GC--------AAGTACTT |
| droMoj3 |
scaffold_6680:3750941-3751063 - |
TTTGTTTTTG---TTT-----------TCTT----TT-----------------------T--GTAGCCTAGGAATTTACC-----TCTTGTGGCATATCAGCTGTTGGC---TTTGGTTGCA-------AACACACG--TTGACTTTTCACACACAGAATTCTG-----TTGAAATGTTTAACACTT |
| droGri2 |
scaffold_14853:566008-566117 + |
CTCTTTTATT----------------------------------GCAATGAGACTGCG-AC--GCA--------TGATTTGGG--CGCAGG-------T---CTATCACCACTTGCACTGACG-------ACTGCTTTTGCTTTTGCT--------ACTACTTTT-----ACTT-GCCAGCGGAACTT |