| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:6979754-6979901 - | GAGTGCCAACAATTTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAATCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGTGATGATCTCATG------AACGATCCTCGGGCAAGCAACACCAACCTG |
| droSim1 | chrX:5492772-5492919 - | GAGTGCCAACAATCTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAACCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGTGATGATCTCATG------AACGATCCTCGGGCAAGCAACACCAACTTG |
| droSec1 | super_24:199934-200081 - | GAGTGCCAACAATCTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAATCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGCGATGATCTCATG------AACGATCCTCGGGCAAGCAACACCAACTTG |
| droYak2 | chrX:14374793-14374940 + | GAGTGCCAACAATCTGGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAATTCGCTGAATAACCTCTGCGGACTGTCGCTGGGCAGCGGCGGTAGCGATGATCTCATG------AACGATCCCCGGGCAAGCAACACCAACCTG |
| droEre2 | scaffold_4690:15890244-15890391 - | GAGTGCCAATAATCTTGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAATTCGCTGAATAACCTCTGCGGACTGTCGCTGGGCAGCGGCGGTAGCGATGATCTCATG------AACGATCCCCGGGCAAGCAACACCAACCTG |
| droAna3 | scaffold_13117:2326296-2326455 - | CAGTGCCAATTCCCTTGGCGGCGGCGGTGGCGGCAACATGTGCAACCTGCCGCCCATGACCAGCAACAATTCGCTTAACAATCTGTGCGGCCTGTCGCTGGGCAGCGGCGGCAGCGACGATCACATGCTT---AATGACCAGCGGCCGAGCAACACCAACCTG |
| dp4 | chrXL_group1a:6391512-6391662 - | CAGTGCCAATAATCTTG------------GCGGCAACATGTGCAACCTGCTGCCAATGACCAGCAACAATTCGCTTAGCAATCTATGCGGACTGTCGCTGGGCAGCGGCGGCAGCGATGATCACATGATGATGCACGATCAGCGATCGAGCAACACCAATCTG |
| droPer1 | super_25:931278-931428 + | CAGTGCCAATAATCTTG------------GCGGCAACATGTGCAACCTGCTGCCAATGACCAGCAACAATTCGCTTAGCAATCTATGCGGACTGTCGCTGGGCAGCGGCGGCAGCGATGATCACATGATGATGCACGATCAGCGATCCAGCAACACCAATCTG |
| droWil1 | scaffold_180777:2984533-2984680 + | CAGTGCCAATTCCCTTGGCG---------GTGGCAATATGTGCAATATGCCACCGATGGCCAGTAACAATTCATTGAACAACCTGTGCGGTCTATCATTGGGCAGCGGCGGCAGCGATGATCTTATG------AACGATCATCGTCCGAGTAATACCAATTTG |
| droVir3 | scaffold_12928:60655-60799 - | CAGCGCCAATTCCCTTG------------GCGGCAACATCTGCAATATGCCGCCAATGGCCAGCAACAATTCGCTGAACAATCTGTGCGGCCTATCGATTGGCAGCGGCGGCAGCGATGATCACATG------AACGATCAGCGCAACAGCAACACCAATTTG |
| droMoj3 | scaffold_6359:1380681-1380818 - | C-----CAATTCCCTCG------------GCGGCAACATCTGCAACATG---CCAATGGCCAGCAACAATTCGCTGAACAATCTGTGCGGCCTATCGATCGGCAGCGGCGGCAGCGATGATCTGATG------AACGATCAGCGCACAAGCAACACCAATTTG |
| droGri2 | scaffold_14853:6692716-6692854 - | CAGCGCCAATTCACTCGGCG---------G---A---CTCTGCAACATG---CCGATGGCCAGCAACAATTCGCTGAACAATCTGTGCGGCCTATCGATAGGCAGCGGGGGCAGCGATGATCTTATG------AACGATCCACGCAACAGCAACACCAATTTG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:6979754-6979901 - | GAGTGCCAACAATTTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAATCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGTGATGATCTCATG---A---ACGATCCTCGGGCAAGCAACACCAACCTG |
| droSim1 | chrX:5492772-5492919 - | GAGTGCCAACAATCTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAACCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGTGATGATCTCATG---A---ACGATCCTCGGGCAAGCAACACCAACTTG |
| droSec1 | super_24:199934-200081 - | GAGTGCCAACAATCTAGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAACTCGCTGAATAATCTCTGCGGATTGTCGCTGGGCAGCGGTGGTAGCGATGATCTCATG---A---ACGATCCTCGGGCAAGCAACACCAACTTG |
| droYak2 | chrX:14374793-14374940 + | GAGTGCCAACAATCTGGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAATTCGCTGAATAACCTCTGCGGACTGTCGCTGGGCAGCGGCGGTAGCGATGATCTCATG---A---ACGATCCCCGGGCAAGCAACACCAACCTG |
| droEre2 | scaffold_4690:15890244-15890391 - | GAGTGCCAATAATCTTGGGG---------GCGGCAACATGTGCCACCTGCCGCCGATGGCCAGCAACAATTCGCTGAATAACCTCTGCGGACTGTCGCTGGGCAGCGGCGGTAGCGATGATCTCATG---A---ACGATCCCCGGGCAAGCAACACCAACCTG |
| droAna3 | scaffold_13117:2326296-2326455 - | CAGTGCCAATTCCCTTGGCGGCGGCGGTGGCGGCAACATGTGCAACCTGCCGCCCATGACCAGCAACAATTCGCTTAACAATCTGTGCGGCCTGTCGCTGGGCAGCGGCGGCAGCGACGATCACATGCTTA---ATGACCAGCGGCCGAGCAACACCAACCTG |
| dp4 | chrXL_group1a:6391512-6391662 - | CAGTGCCAATAATCTTG------------GCGGCAACATGTGCAACCTGCTGCCAATGACCAGCAACAATTCGCTTAGCAATCTATGCGGACTGTCGCTGGGCAGCGGCGGCAGCGATGATCACATGATGATGCACGATCAGCGATCGAGCAACACCAATCTG |
| droPer1 | super_25:931278-931428 + | CAGTGCCAATAATCTTG------------GCGGCAACATGTGCAACCTGCTGCCAATGACCAGCAACAATTCGCTTAGCAATCTATGCGGACTGTCGCTGGGCAGCGGCGGCAGCGATGATCACATGATGATGCACGATCAGCGATCCAGCAACACCAATCTG |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-36.3, p-value=0.009901 |
|---|
 |
| dG=-36.3, p-value=0.009901 |
|---|
 |
| dG=-36.3, p-value=0.009901 |
|---|
 |
| dG=-38.6, p-value=0.009901 |
|---|
 |
| dG=-38.6, p-value=0.009901 |
|---|
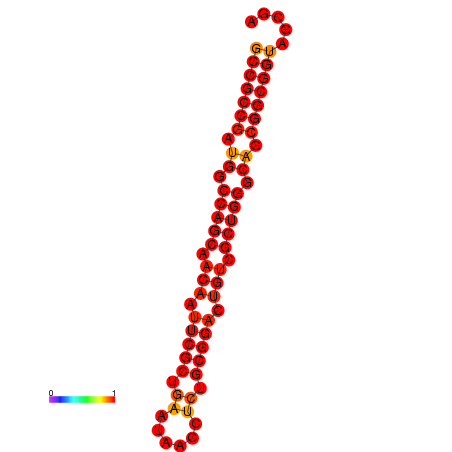 |
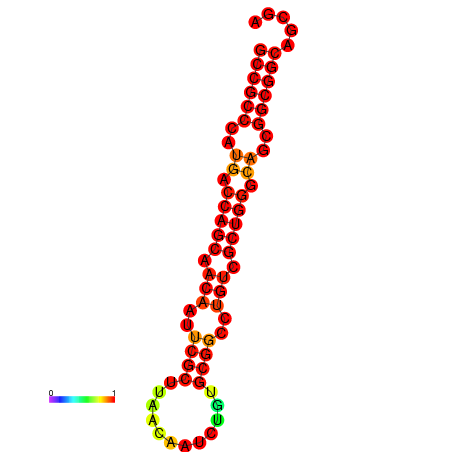
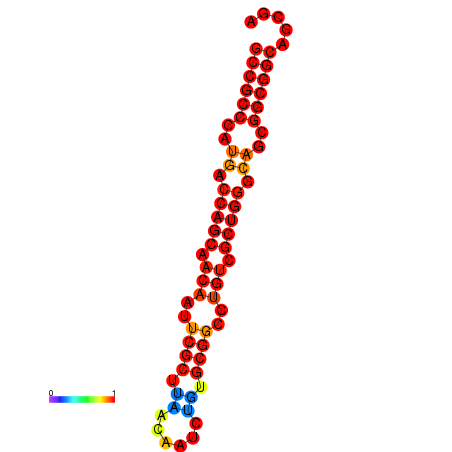
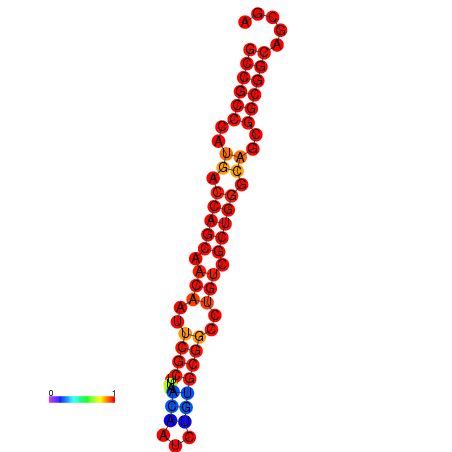
| dG=-30.7, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.0, p-value=0.009901 |
|---|---|---|
 |
 |
 |
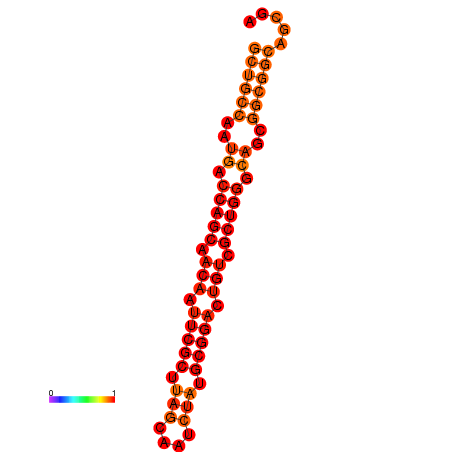
| dG=-33.4, p-value=0.009901 |
|---|
 |
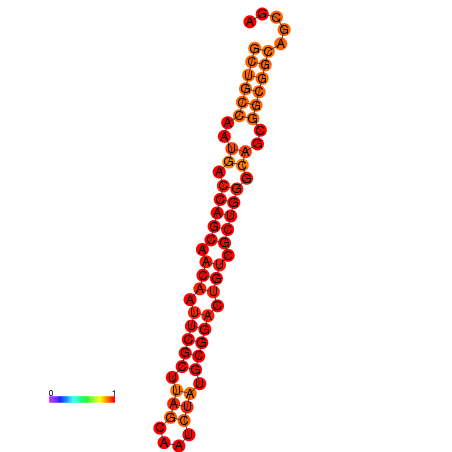
| dG=-33.4, p-value=0.009901 |
|---|
 |
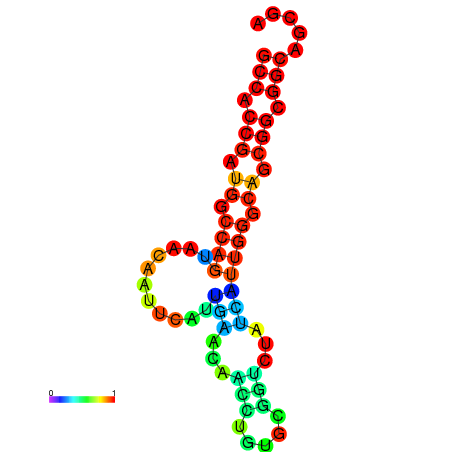
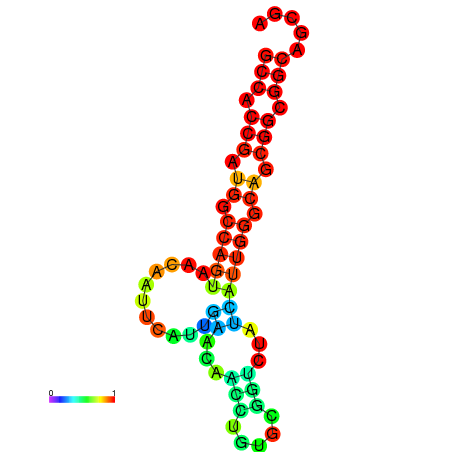
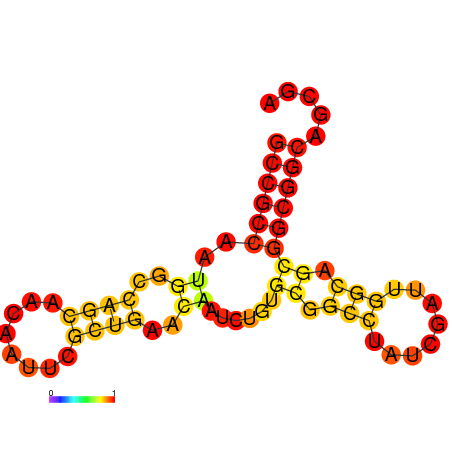
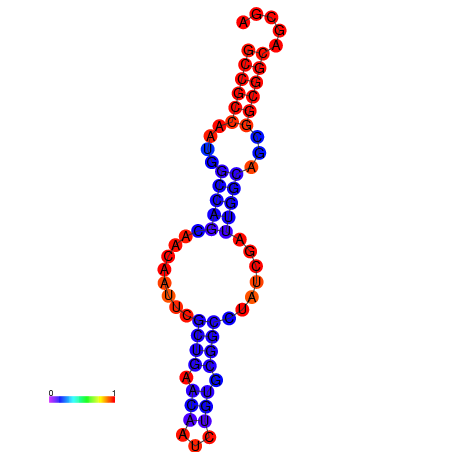
| dG=-19.4, p-value=0.009901 | dG=-19.2, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-19.0, p-value=0.009901 | dG=-18.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
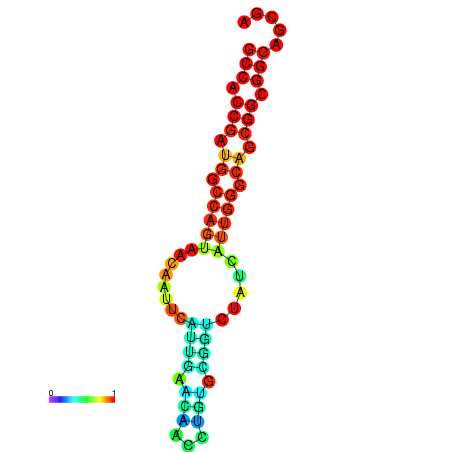 |
 |
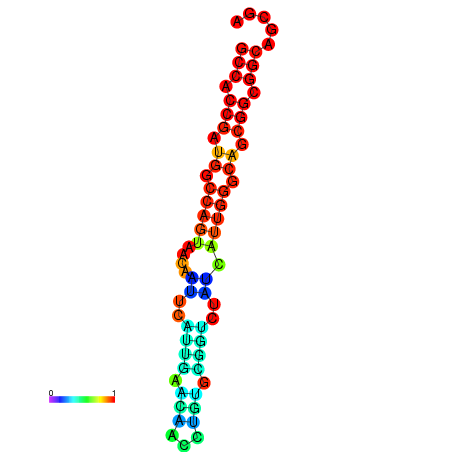
| dG=-25.7, p-value=0.009901 | dG=-23.6, p-value=0.009901 |
|---|---|
 |
 |
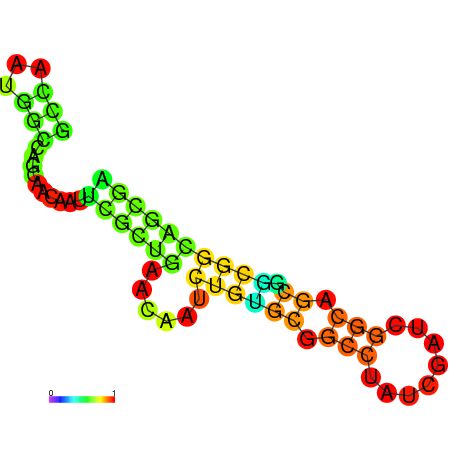
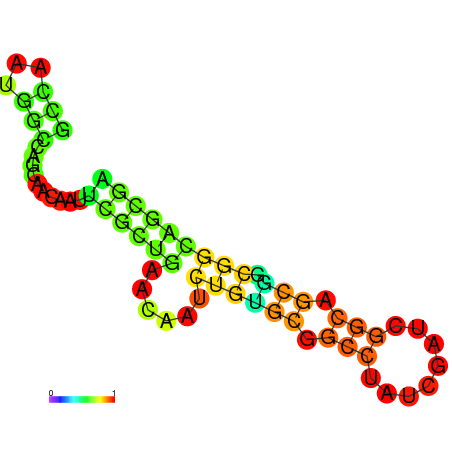
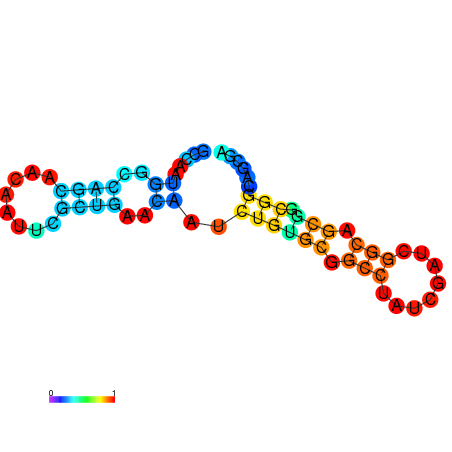
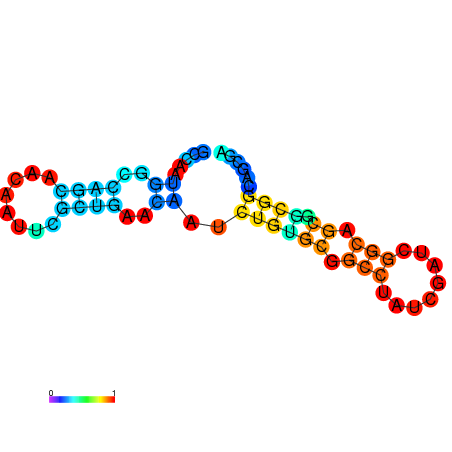
| dG=-18.5, p-value=0.009901 | dG=-18.5, p-value=0.009901 | dG=-17.5, p-value=0.009901 | dG=-17.5, p-value=0.009901 | dG=-16.8, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
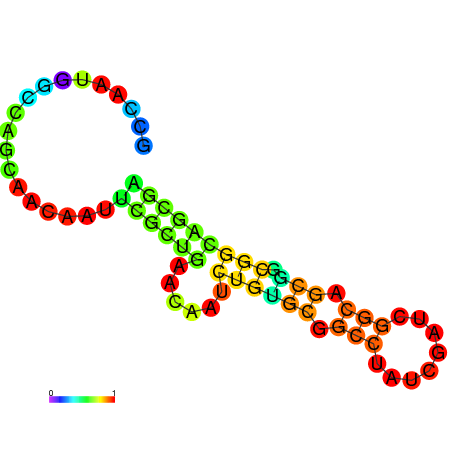 |
| dG=-22.5, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-20.8, p-value=0.009901 |
|---|---|---|
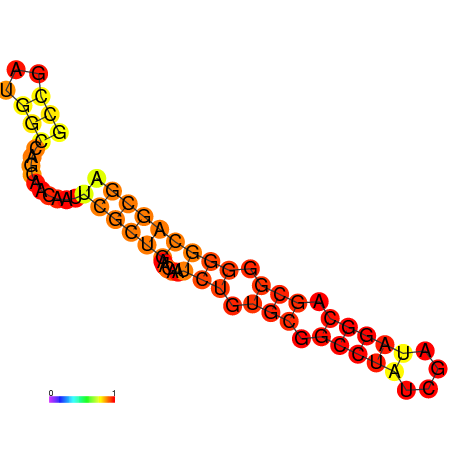 |
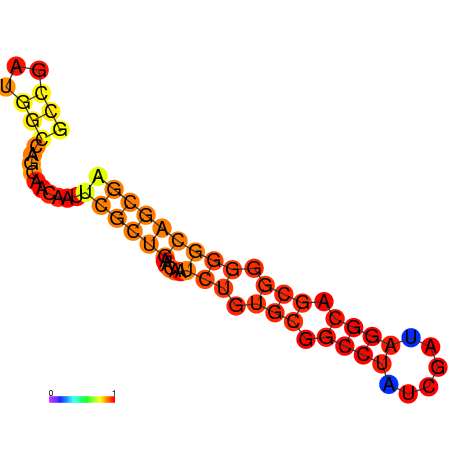 |
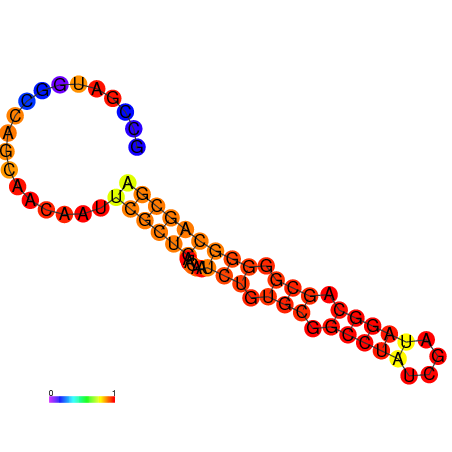 |
Generated: 03/07/2013 at 05:57 PM