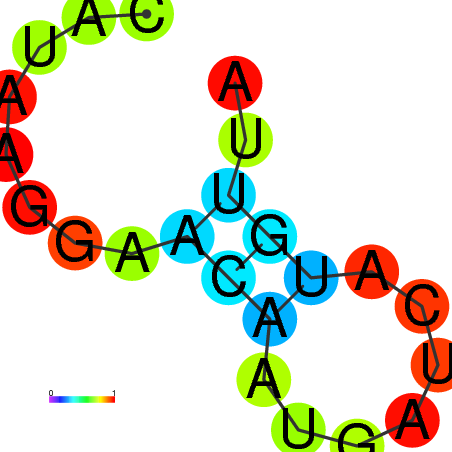
| dm3 |
chrX:1923021-1923142 - |
ATATAAGTGTTCG--------ATTTGGCGCTTGGAATCGATACCCGAGCCATGATAGATTGAAGTCAA-CCCA-ATC----G----------ATC-GC-GGT-TCGAGTGCTCGAGTCTTGTGCGCCGGCATGC-TTGAGAT---GTCCACTA |
| droSim1 |
vgb65a08_5.b1:263-385 - |
ATATAAGTGTTCG--------ATTTGGCGCTTGGAATCGATACCCGAGCCCTGATAGATTGAAGTCAA-CCCA-ATC----G----------ATC-GC-GGT-TCGGGTGCTCGAGTCCTGTGCGCCGGCATGC-TTGAGAT---GTCCACTA |
| droSec1 |
super_10:1708076-1708197 - |
ATATAAGTGTTCG--------ATTTGGCGCTTGGAATCGACACCCGTGCCCTGATAGATTGAAGTCAA-CCCA-ATC----G----------ATC-GC-GGT-TCGGGTGCTCGAGTCTTGTGCGCCGGCATGC-TTGAGAT---GTCCACTA |
| droYak2 |
chrX:6403257-6403382 + |
ACATAAAAGTTCA--------ATTTGGCGCTTGGAATCGTAACTCGAGCCCTCAAAGATTGAAGTCAAATGCA-ATC----G----------ATCCGG-GGTGTCGAGTGCTCGATTCTGGTGCGCCGGCATGCTTTGAGAT---GTCCACTA |
| droEre2 |
scaffold_4644:1895276-1895400 - |
ATATAAGTGTTCG--------ATTTGGCGCTTGGAATCGAACCTCGAGCCCTGACAGATTGAAGTCAA-CCCAAAAC----G----------ATC-CCGGGG-TCGAGTGTTCGAGTCCGGTGCGCCGGCATGCTTTGAGTT---GTCCACTA |
| droAna3 |
scaffold_12929:1917028-1917098 - |
ATATCCTTGACTT-------------------------CATAA-GGAACAATGA---------------------------------------TC-AT--GT--------------TAAGGTTCTCAGGCATGTCTCGAGATTATATCGAATC |
| dp4 |
chrXL_group1a:2597066-2597203 - |
ATATAAATACATACTGCATAGTGTAGCAGAATGGACTCGGAACTCGATCCATGCATGCCACATGCCACATGCC-ACATGTCGCAGAGGGGGCAGC-GT-GGC-TCGAGTGCCAGAGTCCGGCGAG-------GC-ATGAGAC---TACTTACC |
| droPer1 |
super_28:1034311-1034357 - |
A--------------------------------------------------------------------------------------------GC-GT-GGC-TCGAGTGCCAGAGTCCGGCGAG-------GC-ATGAGAC---TACTTACC |
| droWil1 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6473:2911987-2912050 + |
ATATATGTGTGTG--------TGTTTGTGTATTGTATTTAGATTTGAGCTATAAAATGTTTAATAAAA-CTCA-------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
--------------------------------------------------------------------------------------------------------------------------------------------------------- |