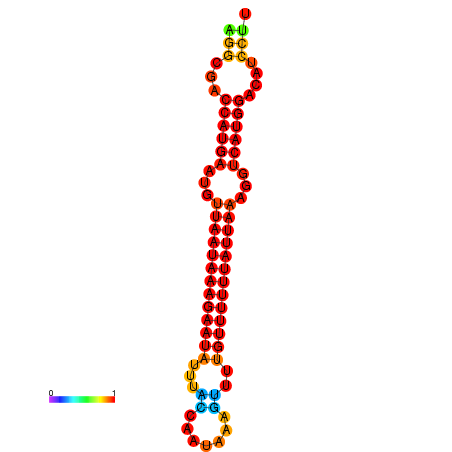
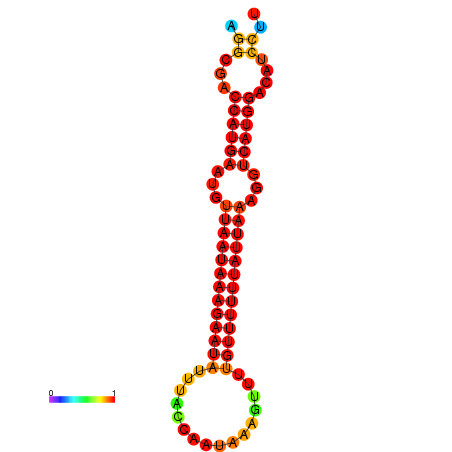
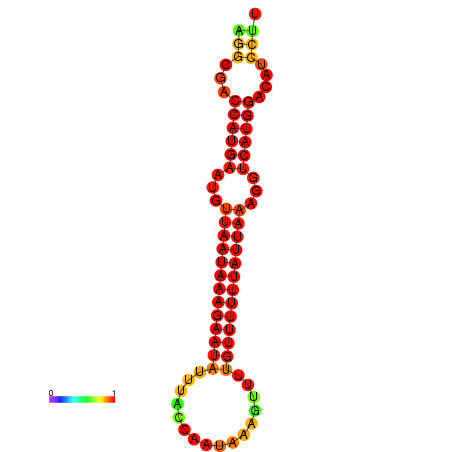
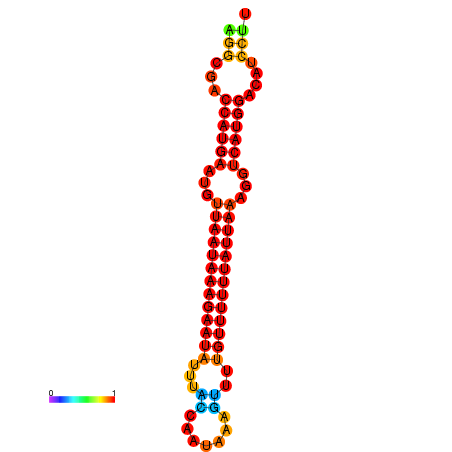
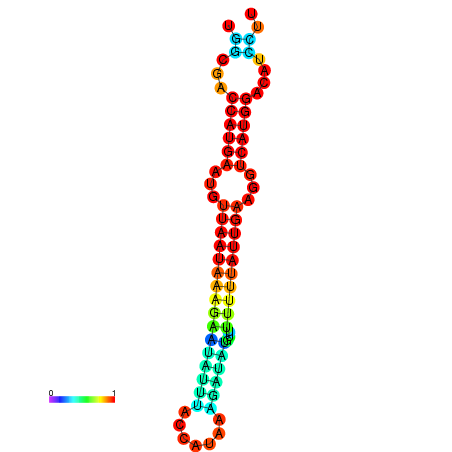
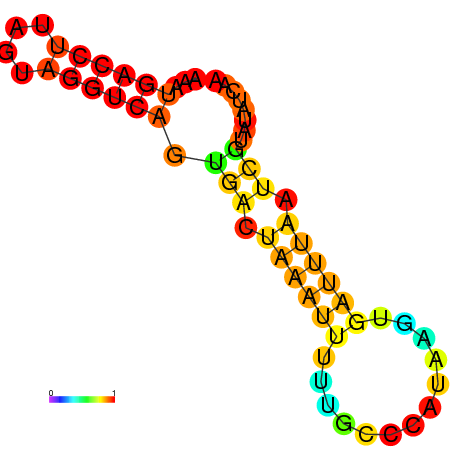
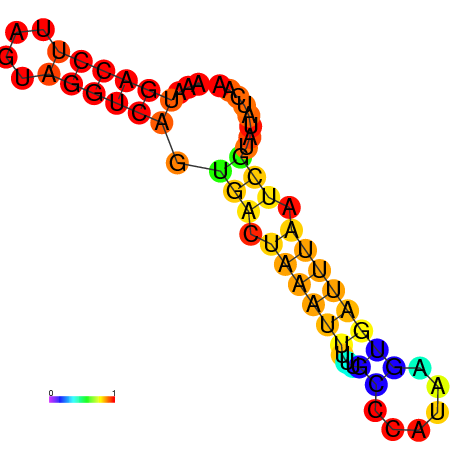
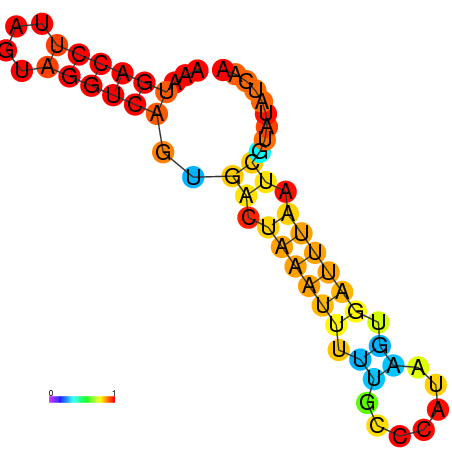
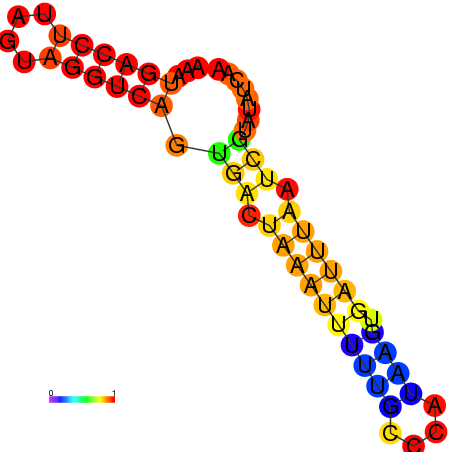
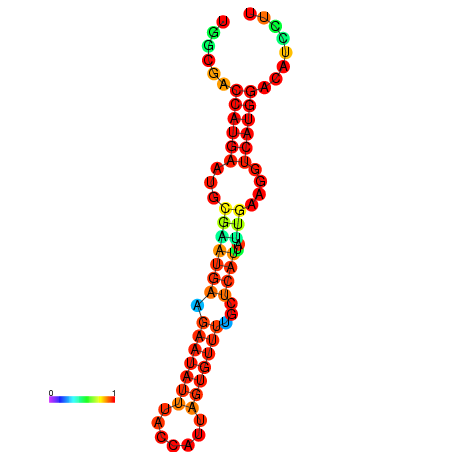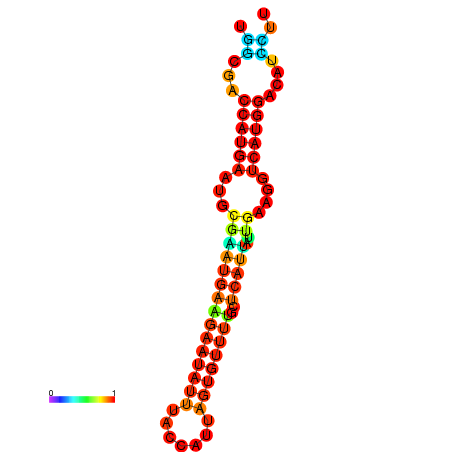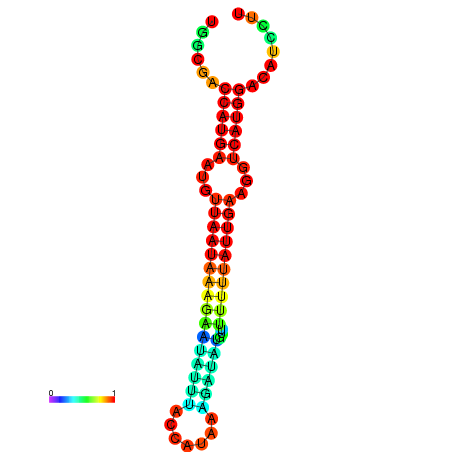| dm3 |
chr3R:18599846-18599969 + |
CGTAAGAATA-------------TTG-------CACTAAAGTTCACAGGCGACCATGA-AT---GTTAATAAAG----A--------ATAT---------------------TTACCAATAAAGTTTTATTTTTTATTGAAGGTCATGGACATCCTTGAACCGATTTTGCAACATG-----CATGT |
| droSim1 |
chr3R:18412164-18412275 + |
CGTAAAAATA-------------TTG-------CACGAAAGTTCGCAGGCGACCATGA-AT---GTTAATAAAG----A--------ATAT---------------------TTACCAATAAAGTTTTGTTTTTTATTAAAGGTCATGGACATCCTTGAACG--------------------GTGT |
| droSec1 |
super_0:18960705-18960813 + |
CGTAAAAATA-------------TTG-------CACGAAAGTTCGCAGGCGACCATGA-AT---GTTAATAAAG----A--------ATAT---------------------TTACCAATAAAGTTTTGTTTTTTATTAAAGGTCATGGACATCCTTGAACG--------------------GTGT |
| droYak2 |
chr3R:19450585-19450700 + |
TGCTCAAATT-------------TTG-------CACGAAAGTTCGATGGCGACCATGA-AT---GCGAATGAAG----A--------ATAT---------------------TTACCATTAGTGTTTTGCTCATTATTGAAGGTCATGGACATCCTTGA-----TTTTGCAAAATG--------TT |
| droEre2 |
scaffold_4820:9473651-9473779 - |
CGCTAAAATA-------------TGG-------CACGAAAGTTCACTGGCGACCATGA-AT---GTTAATAAAG----A--------ATAT---------------------TTACCATAAAGATATGTTTTTTTATTGAAGGTCATGGACATCCTTGAACCGATTTTGCAATATGTTCTACGTGT |
| droAna3 |
scaffold_13340:12717426-12717507 - |
GGGGAAAAAA-------------TGA-------AAAGAA-----ACAAATGACCTTAGTAGGTCAGTGACTAAATTTTTGCCCATAA----GT-----------------------------GATTTAATCGTATATCAA---------------------------------------------- |
| dp4 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droPer1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droWil1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droVir3 |
scaffold_12822:414561-414651 - |
TGCAAGCAAA-------------TTT-------TGCCTAAGTTCTGA------------------CTAATAACT----A--------ATGT---------------------ATGTACTTAATGTTATTGTGATAAATTAATGTTATTTGTAACCAA------------------------AATGT |
| droMoj3 |
scaffold_6540:33042485-33042630 - |
CGTAGTTTGACCACATAGAGACATCAAGTCTAAGAGGAAAACTCA--------TTTGG-AC---ACTGATTAAAAATCAGTCCCGAT----GAGAGCGAT------------AT---ATTAACATTAATTTTCTTATTGCATTTTACGTATATTTTTTTTTTGCTTTTAACA---------TACAT |
| droGri2 |
scaffold_15074:3394740-3394844 + |
GTATGATACA-------------------------TTAAAGTTCTCCATAATCAATTT-GT---GT-------------------------GTGTGTGTGTCATCGACAAATTTACCATCTTAACTACACTTATTTTCAAAAT-------------------A-TTTCACATA--------TATTT |