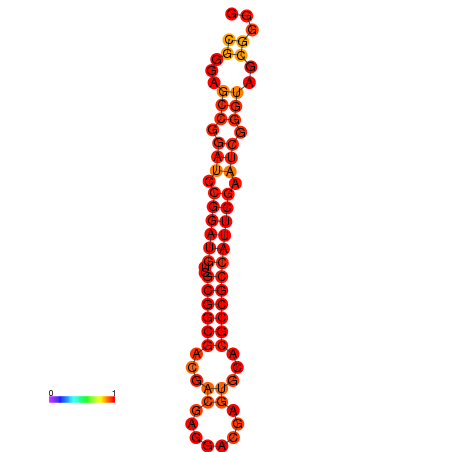
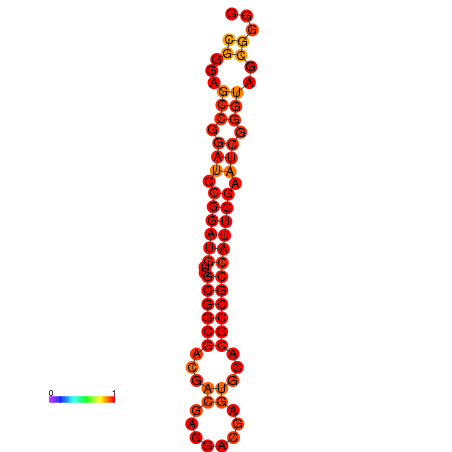
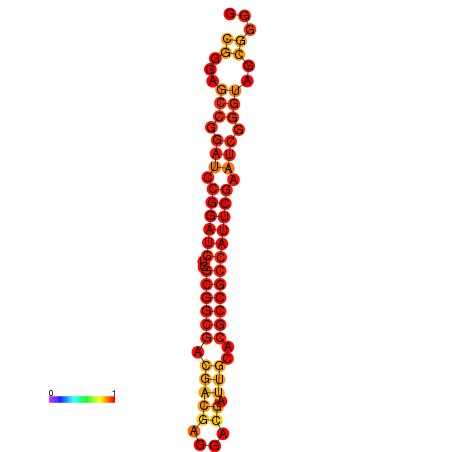
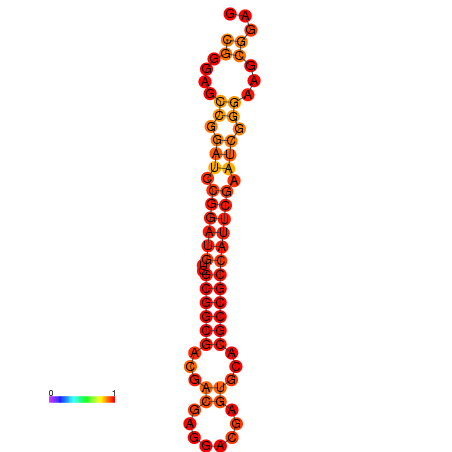
| dm3 |
chr3R:18306519-18306675 + |
CGCCGGCGAGTGGTTATCCGGGTGCGCTGGACGATCTGATCTTCTCGGGAGCCGGATCCGGATGTCGCGGCGACGACGAGGACGAGTGCACGCCGCCATTCGAATCGGGTAGCGGGGATGATCTCATAACACCGGTCTACGTGCCGCCCACCAAGCA |
| droSim1 |
chr3R:18115675-18115831 + |
CGCCGGCGAGTGGTTATCCGGGTGCGCTGGACGATCTGATCTTCTCGGGAGCCGGATCCGGATGTCGCGGCGACGACGAGGACGAGTGCACGCCGCCATTCGAATCGGGTAGCGGGGATGATCTCATAACACCGGTCTACGTGCCGCCCACCAAGCA |
| droSec1 |
super_0:18660821-18660977 + |
CGCCGGCGAGTGGTTATCCGGGTGCGCTGGACGATCTGATATTCTCGGGAGCCGGATCCGGATGTCGCGGCGACGACGAGGACGATTGCACGCCGCCATTCGAATCGGGTAGCGGGGATGATCTCATAACACCAGTCTACGTGCCGCCCACCAAGCA |
| droYak2 |
chr3R:19151970-19152126 + |
CGCCGGCGAGTGGTTATCCGGGTGCGCTGGACGATCTGATCTTCTCGGGAGCCGGATCCGGATGTCGCGGCGACGACGAGGACGAGTGCACGCCGCCATTCGAATCGGGAAGCGGAGATGATCTCATAACACCGGTCTACGTGCCGCCCACCAAGCA |
| droEre2 |
scaffold_4820:9770831-9770987 - |
CGCCGGCGAGTGGTTATCCGGGTGCGCTGGACGATCTGATCTTCTCGGGAGCCGGATCCGGATGTCGCGGCGACGACGAGGACGAGTGCACGCCGCCATTCGAATCGGGAAGCGGGGATGATCTCATAACACCGGTCTACGTGCCGCCCACCAAGCA |
| droAna3 |
scaffold_12911:5347592-5347748 + |
CTCCGGCGAGTGGCTATCCGGGTGCCCTGGACGATCTGATCTTCTCGGGAGCGGGATCCGGGTGCCGGGGCGATGACGAGGACGAGTGCACGCCACCCTTCGAGTCGGGCAGCGGGGACGACCTGATCACTCCGGTGTATGTACCGCCCACCAAGCA |
| dp4 |
chr2:13773155-13773311 - |
CGCCGGCCAGCGGCTATCCCGGCACCATTGACGATCTGATCTTCTCCGGGGCCGGGTCCGGGTGCCGCGGCGACGACGAGGACGAGTGCACACCGCCCTTCGAGTCGGGCAGCGGCGATGACCTCATCACGCCCGTGTATGTGCCACCCACCAAGCA |
| droPer1 |
super_0:6673715-6673871 + |
CGCCGGCCAGCGGCTATCCCGGCACCATTGACGATCTGATCTTCTCCGGGGCCGGGTCCGGGTGCCGCGGCGACGACGAGGACGAGTGCACACCGCCCTTCGAGTCGGGCAGCGGCGATGACCTCATCACGCCCGTGTATGTGCCACCCACCAAGCA |
| droWil1 |
scaffold_181089:2782020-2782176 + |
CGCCGGCCAGTGGCTATCCTGGGGCCATAGATGATCTTATATTCTCCGGTGCCGGATCCGGTTGTCGCGGCGACGATGAAGACGAATGTACGCCGCCCTTCGAGTCGGGCAGTGGAGATGATCTAATCACACCTGTCTATGTGCCTCCGACAAAACA |
| droVir3 |
scaffold_12822:645540-645696 - |
CGCCTGCGAGCGGCTATCCGGGCGCCATAGACGATCTGATCTTCTCCGGGGCCGGGTCCGGTTGTCGCGGCGACGACGAGGACGAATGCACGCCCCCCTTCGAGTCGGGCAGCGGGGATGACCTAATAACGCCCGTCTATGTGCCGCCCACCAAGCA |
| droMoj3 |
scaffold_6540:33300209-33300365 - |
CGCCGGCTAGTGGCTATCCGGGCGCCATAGACGACCTGATCTTCTCCGGAGCCGGATCTGGGTGTCGCGGCGACGACGAGGATGAATGCACGCCGCCCTTCGAGTCGGGCAGTGGGGATGACCTGATCACACCCGTCTATGTGCCGCCCACCAAACA |
| droGri2 |
scaffold_15074:3142694-3142850 + |
CGCCGGCGAGTGGTTATCCGGGTGCGATAGACGATCTGATCTTCTCCGGTGCGGGATCCGGGTGTCGCGGCGACGATGAGGACGAATGCACGCCGCCCTTCGAGTCGGGCAGCGGGGATGATCTAATTACGCCCGTCTATGTGCCACCCACCAAACA |