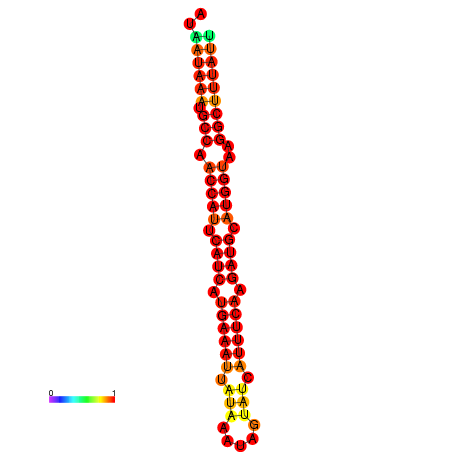
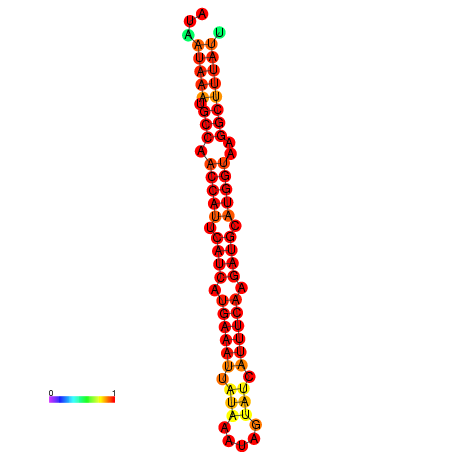
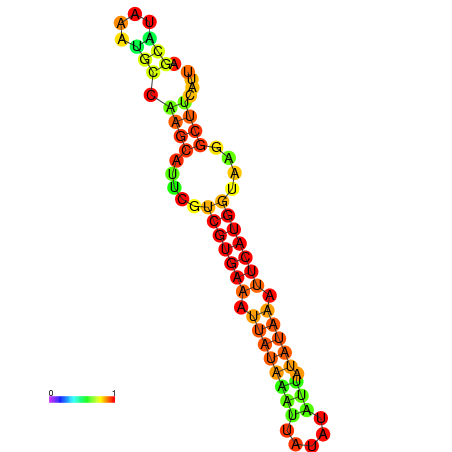
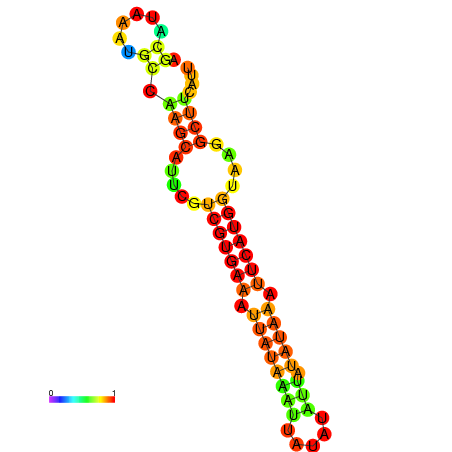
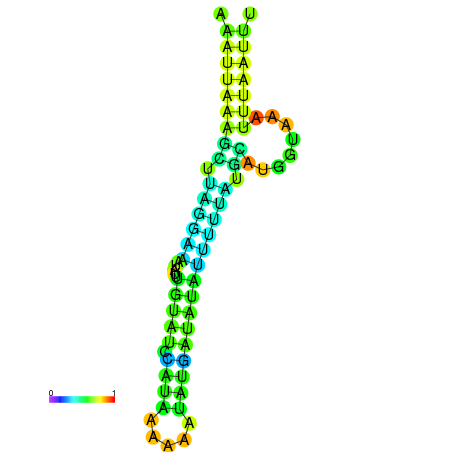
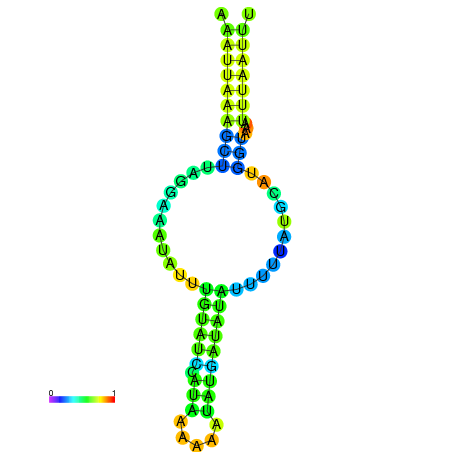
| dm3 |
chr3R:3277158-3277280 + |
TTTATACATTTG--TTTAT-----------------------AATGCACATAATAAATGCCAACC------ATTCATCATGA-AATTATAAATAGTAT--CATTTCAAGATGCATGG-TAAG-GCTT------TATTTT---------------------------------------------G--------TTTTGCA---ATCAAAGACATCTTTCTT |
| droSim1 |
chr3R:3321450-3321569 + |
TTTATATATATT-ATTTAT-----------------------TATGCTCAGCATAAATGCCAAGC------ATTCGTCGTGA-AATTATAAATTATATATTA--TATAAATTCATGG-TAAG-GCTT------CATTTT---------------------------------------------T--------TAACGCA---GTCTAAGACATC----TT |
| droSec1 |
super_6:3368064-3368189 + |
TTTATATATATT-ATTTAT-----------------------TATGCTCAGCATAAATGCCAAGC------ATTCGTCGTGA-AATTATAAATTATATATTATTTCTAGATTCATGG-TAAA-GTTT------CATTTT---------------------------------------------G--------TAACGCA---GTCTAAGACATCTTTCTC |
| droYak2 |
chr2L_random:3663454-3663574 - |
TTTAGCCAAGTT-A-ATT--------------------------TGGTAAAAACAAAAACCAGAA------ACCGGCTACAC-GATGAAACGCTAAATGCCATTTTAGCAGTC--AG-GTAATTATA------TATT--------------------------------------------------T--AT-TTTTAAA---TTTAAACCCATTTTTTAT |
| droEre2 |
scaffold_4784:2237866-2237984 - |
TTTCCAAATTTG-A-ATGC---AGA-TTTTT------------------AGACAATTGCTCCAGA------AACATT------------TAGAGGTATGCCACTTCAAAATACGA----TAATTGAAGGTTGAATTATG---------------------------------------------G-----AC--TTTAG----AACTTCAGCCTTCTTTTT |
| droAna3 |
scaffold_13340:2198726-2198841 + |
TTAATAATTTTA--TTTAT-----------------------CGAAC------TAATT-AGGCTT------TTCATACAGATTTAAGCTTAATATTAAATTA--TAG-------------------------------------------------------AGCACTGGCTGCGGTGGTATATA--T--AT--TTTTC----AATAAACACATTTTTATG |
| dp4 |
chrXR_group6:912830-912970 - |
TTTATACCACGC--ACTAGATTGGGTTCTCCAGATAACGAATGTTGTCCAGTAAAATCCCAATCCATTCAAATGGATTATGT-GACCA-------------GTGTTAAGATGCGTCG-AGGG-CAGT------CAGCTT---------------------------------------------G--------TGTTGCT---ATCTCGCTCCCCTCCGCC |
| droPer1 |
super_3:249021-249147 + |
TTTTAAATCTGTTCCCTAAAATATACTTTTTGAAC------------------GAG-------------------------A---------------------------------------G-AGTA------TATATGTATGTATGCCGAGCAACTTCTAGTACCCCCTACTCAAT-A----TGTTTTTTTTTGGCTTAGTAAAATAATACATCTATATC |
| droWil1 |
scaffold_180708:152570-152690 - |
TTTATATATATA-T-ATAT---GAT-GTAGA---TATA---TATTTTTTAT----ATTTAAATTTTT----------------------TTAAAGTGA--TTTTTGTCAA-GGCTAG-AAAA-GCTT------TATTTT-------------------------------------------TTGTTT-----TGTTTTA---GTTTATGCGATATTTCTT |
| droVir3 |
scaffold_135:841-956 - |
TATATATATGTG-A-ACAT---AAC-TTATA-------------------TAATA------------------------------------CATATGA--CATACTGCCAGAC--AAATAAA-CGTA------ACTATTTATGTTTGAA-------ATCAAGATCACCAAAATAAATGATGATTT--T--AT--TT--------------------TACAA |
| droMoj3 |
scaffold_6308:1934837-1934961 - |
TTTATATATTAT-A-AAAA---AAA-T-----------------TACTTAAATTAAAGCTTAGGA------AATATTTGTAT-CCATAAAAAATATGA--TATATTTTTATGCATGG-TAAA-TTTA------ATTTTT---------------------------------------------A--------ATTTCTA---ATAAAAAAAATTATATAT |
| droGri2 |
scaffold_14906:7885716-7885837 + |
ATTATTATTTTT--TTTTT-----------------------A--TTTCAAGTTAATTTACAATT------ATCGAACCGGT-TAAACTAAATTTTAGCTTA--TAGATATTTATTG--------------------------------------------------------CTTAATTAACTGTTTTT---TTTTGTT----GTTAAAACATTTTTTTT |