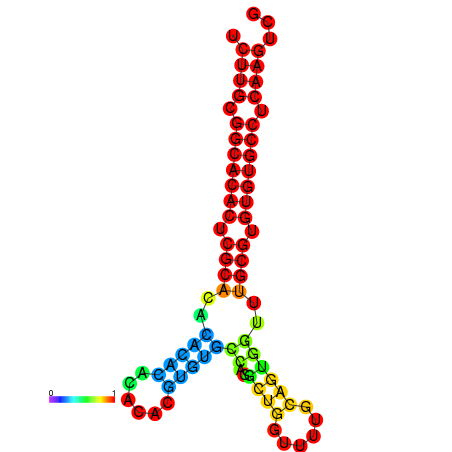
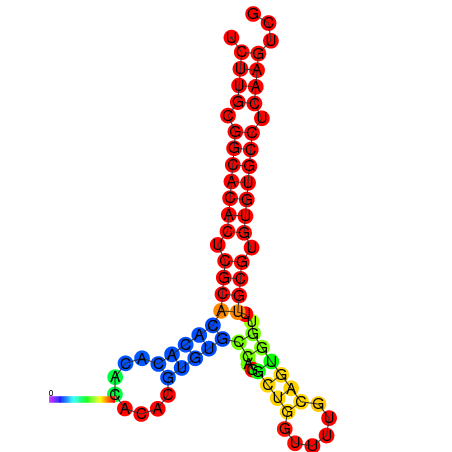
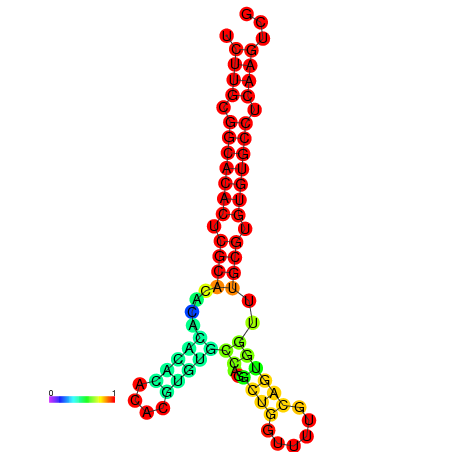
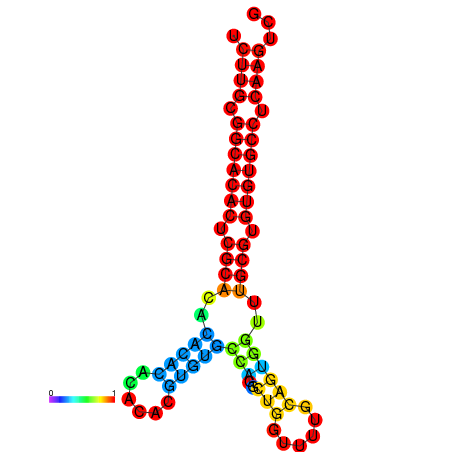
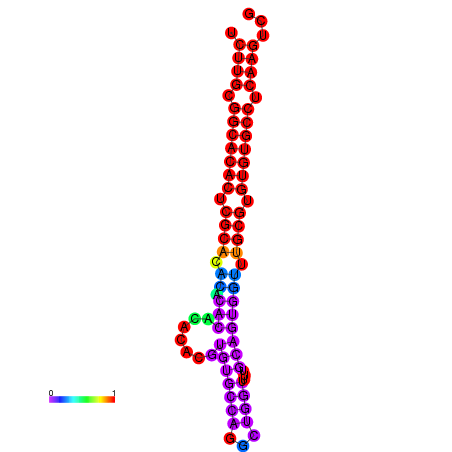
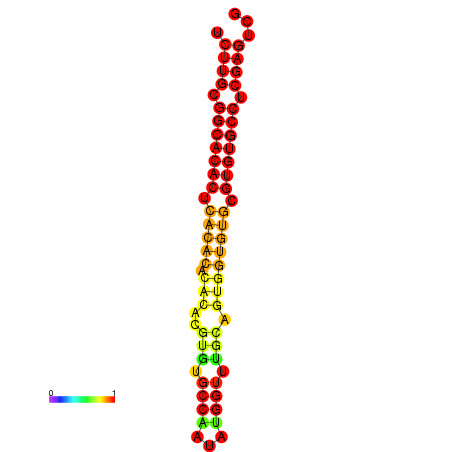
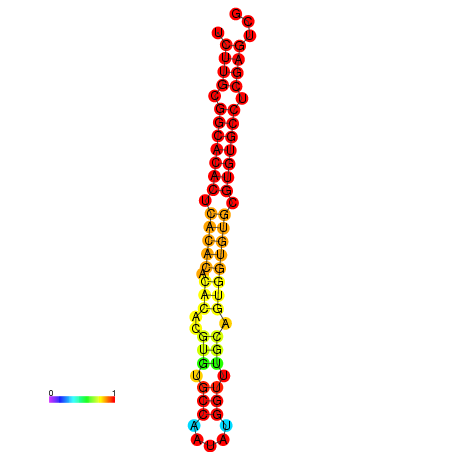
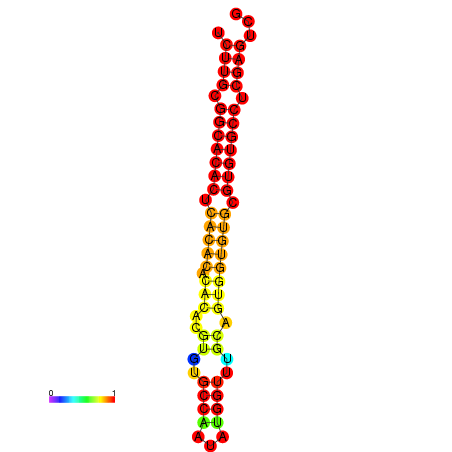
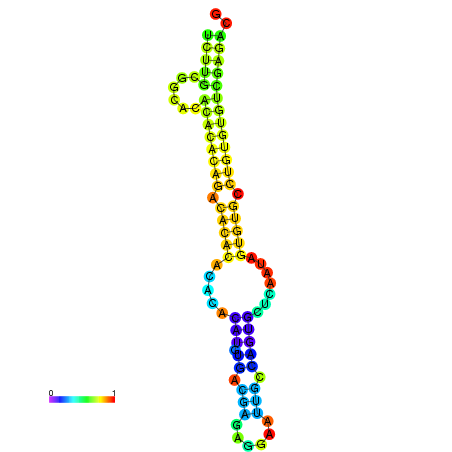
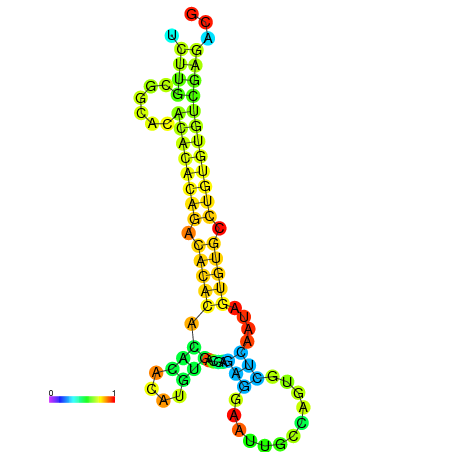
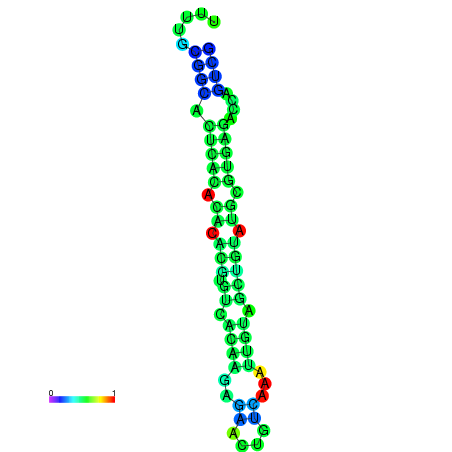
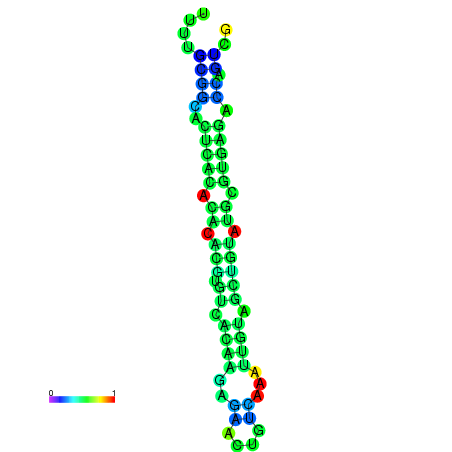
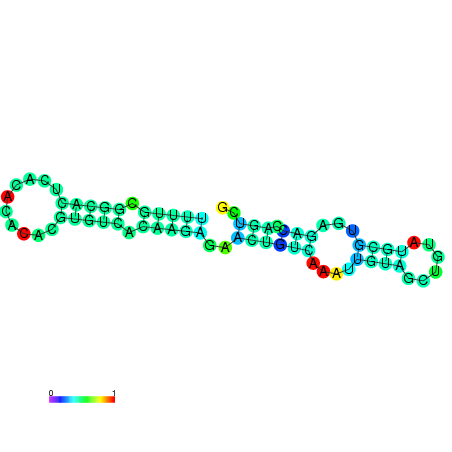
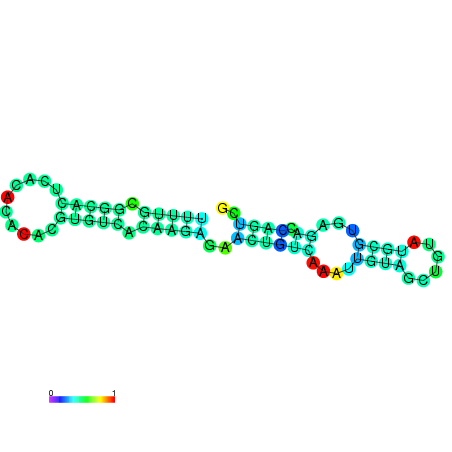
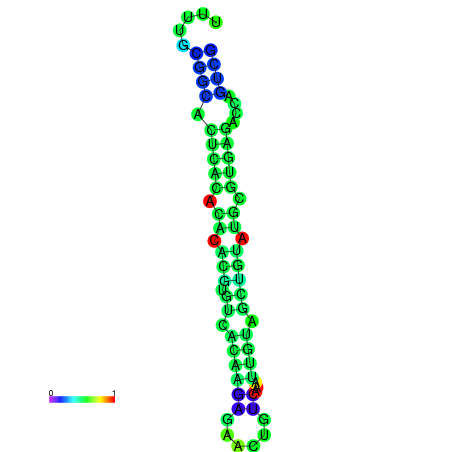
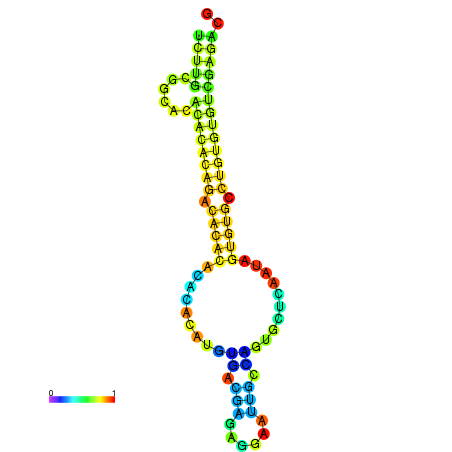| dm3 |
chr3R:27044980-27045109 - |
TGCGAAACG----------------TGTTTGA-T------AATGCGCCGCCATCTTGCGGCACACTCGCACA-C------ACACA-CGTGTGCC-----------ATTCTGG-------TTTTGC--AGTGGTGTGCGTGTGTGCCTC-------GAGTCGTTGCGTCACTGGAAAAGTT-----------AGAAAATC |
| droSim1 |
chr3R:26688146-26688275 - |
TGCGAAACG----------------TGTTTGA-T------AATGCGCCGCCATCTTGCGGCACACTCGCACA-C------ACACA-CGTGTGCC-----------AGGCTGG-------TTTTGC--AGTGGTGTGCGTGTGTGCCTC-------GAGTCGTTGCGTCACTGGAAAAGTT-----------AGAAAATC |
| droSec1 |
super_4:5862973-5863106 - |
TGCGAAACG----------------TGTTTGA-T------AATGCGCCGCCATCTTGCGGCACACTCGC---ACACACACACACA-CGTGTGCC-----------AGGCTGG-------TTTTGC--AGTGGTTTGCGTGTGTGCCTC-------AAGTCGTTGCGTCACTGAAAAAGTT-----------AGAAAATC |
| droYak2 |
chr3R:27981306-27981430 - |
TGCGAAACG----------------TGTTTGA-A------AATCCGTCGCCATCTTGCGGCACACTCAC---AC------ACACA-CGTGTGCC-----------AATATGG-------T-TTGC--AGTGGTGTGCGTGT--GCCTC-------GAGTCGTTGCGTCACTGGAAAAGTC-----------AGAAAATC |
| droEre2 |
scaffold_4820:935924-936048 + |
TGCAAAACG----------------TGTTGGA-A------AATTCGCCGCCATCTTGCGGCACACACA-----C------ACACA-CGTGTGCC-----------AGTCTGG-------T-TTGC--AGTGGTGTGCGTGTGGGCCTC-------GAGTCGTTGCGTCACTGGAAAAGTC-----------CGAAAATC |
| droAna3 |
scaffold_13340:14436299-14436405 - |
CTCAAAACG----------------TGTTCAA-TGG----A----ATCGCCATCTTGCGGCTCACAC---------------ACA-CATGTGAC---------------------------------AGTTGTTCGTGCGTGTGTCTGG------GAGTCGTTGCGTCACTGAGAAAGTC-----------GGAAAAT- |
| dp4 |
chr2:85298-85449 + |
TAC-------ATTTTCTCGAAAACGTGTCGAA------------TGTCGCCATCTTGCGGCACACACAC---AGACACACACACA-CATGTGAC-----------GAGAGGAATTGCCAGT--GCTCAATAGTGTGCCTGTGTG--TC-------GAGACGTTGCGTCACTGAAAAAGTCGAACAGC--AAAGTAAATT |
| droPer1 |
super_7:2877006-2877157 + |
TAC-------ATTTTCTCGAAAACGTGTCGAA------------TGTCGCCATCTTGCGGCACACACAC---AGACACACACACA-CATGTGAC-----------GAGAGGAATTGCCAGT--GCTCAATAGTGTGCCTGTGTG--TC-------GAGACGTTGCGTCACTGAAAAAGTCGAACAGC--AAAGTAAATT |
| droWil1 |
scaffold_181108:712012-712143 - |
TGAAAAATGTATTTTATC--TA--------GA-G------ATATTGTCGCCATTTTGCGGCACTCAC-------------ACACA-CGTGTCAC-----------AAGA-GAACTGTCAAATTGT--AGCTGTATGCGTGAG----AC-------CAGTCGTTGCGTCACAGCAAAAGTT-----------TTCAAATT |
| droVir3 |
scaffold_12855:1227223-1227391 + |
TAA------TATTTTTTC--AAACGTGTTCAG------------TGTCGCCATCTTGCGGCACATACAC---ACACATTCAGACAAGATGTCACACGGACGTACGAAGA-GAACTGTCAGT--GTGTT-TTGTGTGCCGTAGTG--TGGAGTGTAAATTTGTTGCGTCACTAGAAAAGTTCAACTGT-TGAAGGAAATC |
| droMoj3 |
scaffold_6540:26974787-26974936 + |
TAA------CATTTTTTC--AAACGTGTTCAA------------TGTCGCCATCTTGCGGCTCACGCAC---ACACTCA--TGCAAGATGTCACACGGGCGTATGAAGA-GAACTGTCAGT--GTT--AC------------TGTGTG-------AAGTCGTTGCGTCACTAGAAAAGTTCAACGATTTGAAGAAAATT |
| droGri2 |
scaffold_15245:5068344-5068468 + |
G-CGACAAG----------------TGGTTGACTGCACCTGCACCCTCC-CAA---CAGACACACTCACACA-C------ACACA-CACAACCC-----------TCTGTGT-------GTGTGC--AAGTGTGTGTGTGTGTGTGTC-------AAA-CGCAGCA--ACTGCTGAAGTT-----------T----AAC |