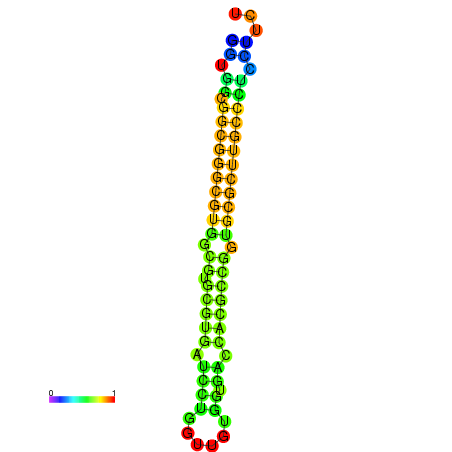
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:26876804-26876940 - | GCACTCCCAGTCCCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGCGTGATCCTGGTTGTGGTGACCACGCCGGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGCT---------------- |
| droSim1 | chr3R:26523820-26523956 - | GCACTCCCAGTCGCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGGGTGATCCTGGTTGTGGTGACCACGCCCGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGTT---------------- |
| droSec1 | super_4:5701489-5701625 - | GCACTCCCAGTCGCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGTGTGATCCTGGTTGTGGTGACCACGCCGGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGCT---------------- |
| droYak2 | chr3R:27815996-27816132 - | GCACTCCCAATCCCGGTCGTAGATGTGGGCGTACGTGGCGGCAGGCGTGGCATGGGTGATCCTGGTGGTGGTGACCACTCCAGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTTTGGACCCTACCTTGCT---------------- |
| droEre2 | scaffold_4820:1105506-1105642 + | GCACTCCCAATCCCGGTCGTAGATGTGGGCGTACGTGGCGGCAGGCGTGGCATGGGTGATCCTGGTTGTGGTGACCACTCCAGTACGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTTTGGACCCTGCCCTGCT---------------- |
| droAna3 | scaffold_12911:4046748-4046884 + | GCACTCCCAATCCCGATCGTAGATGTGGGCGTAGGTTGCAGCTGGCGTGGCGTGGGTAATCCGAGTGGTGGTCACTATTCCAGTTCGCTTCTTCTCCTTTTGGGCCCACTCCATGACGCTCTGAACGCGTCCTTGCT---------------- |
| dp4 | chr2:271882-272018 + | GCACTCCCAGTCGCGATCGTAGATGTGGGCGTAGGTGGCCGCAGGCGTGGCATGCGTAATCCTCGTTGTGGTCACGATTCCCGTGCGCTTGCCTTCCTTCTGGGCCCACTCCATGATAGTCTCAATCCGGCCCAGCT---------------- |
| droPer1 | super_7:3056347-3056483 + | GCACTCCCAGTCGCGATCGTAGATGTGGGCGTAGGTGGCCGCAGGCGTGGCATGCGTAATCCTCGTTGTGGTCACGATTCCCGTGCGCTTGCCTTCCTTCTGGGCCCACTCCATGATAGTCTCAATCCGGCCCAGCT---------------- |
| droWil1 | scaffold_181108:531799-531935 - | GCATTCCCAGTCCCGATCATATATATGCGCATAGGTTGAGGCAGGCGTGGCATGTGTAATTCTGGTAGTGGTAACAATTCCCGTTCGTTTACCCGCACGCTGGGCCCAATCCATAATGGATTCCACCCGACCCTGTT---------------- |
| droVir3 | scaffold_12855:1470098-1470234 + | GCACTCCCAGTCCCGATGATAGACACGCGCAAAGGTGGCTGCGGGCGTGGCGTGCGTAATGCGTGTGGTGGTCACCACGCCAGTGCGTTTGCCTTCACGCTGTGCCCAGTCCATGATGGAGTCGAGTCGACCCTGGC---------------- |
| droMoj3 | scaffold_6540:27213622-27213758 + | GCACTCCCAGTCCCGATGATAGATCCTTGCATAGGTGGCCGCAGGTGTGGCGTGTGTTATACGCGTGGTAGTCACGACGCCGGTCCGCTTGCCCTCACGCTGTGCCCAGTCCATGATGGAGTCGAGTCGACCCTGGC---------------- |
| droGri2 | scaffold_15203:6653367-6653519 - | GCACTCCCACCGCCGATCGGCGACATGGGCGTAGAGGGCAGCGGGTGTGGCATGGGTTACCCGCGTTGTGGTCACAAATCCAGTGCGCATGCCATCCACCTGCGCCCAGTTGAATATGTTCTGGACGTGATGTTCCTCCTTGAGAGAGGCACT |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:26876804-26876940 - | GCACTCCCAGTCCCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGCGTGATCCTGGTTGTGGTGACCACGCCGGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGCT |
| droSim1 | chr3R:26523820-26523956 - | GCACTCCCAGTCGCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGGGTGATCCTGGTTGTGGTGACCACGCCCGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGTT |
| droSec1 | super_4:5701489-5701625 - | GCACTCCCAGTCGCGGTCGTAGATGTGGGCGTAGGTGGCGGCGGGCGTGGCGTGTGTGATCCTGGTTGTGGTGACCACGCCGGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTCTGGACCCTGCCTTGCT |
| droYak2 | chr3R:27815996-27816132 - | GCACTCCCAATCCCGGTCGTAGATGTGGGCGTACGTGGCGGCAGGCGTGGCATGGGTGATCCTGGTGGTGGTGACCACTCCAGTGCGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTTTGGACCCTACCTTGCT |
| droEre2 | scaffold_4820:1105506-1105642 + | GCACTCCCAATCCCGGTCGTAGATGTGGGCGTACGTGGCGGCAGGCGTGGCATGGGTGATCCTGGTTGTGGTGACCACTCCAGTACGCTTGCCCTCCTTCTGGGCCCACTCCATGACGCTTTGGACCCTGCCCTGCT |
| droAna3 | scaffold_12911:4046748-4046884 + | GCACTCCCAATCCCGATCGTAGATGTGGGCGTAGGTTGCAGCTGGCGTGGCGTGGGTAATCCGAGTGGTGGTCACTATTCCAGTTCGCTTCTTCTCCTTTTGGGCCCACTCCATGACGCTCTGAACGCGTCCTTGCT |
| dp4 | chr2:271882-272018 + | GCACTCCCAGTCGCGATCGTAGATGTGGGCGTAGGTGGCCGCAGGCGTGGCATGCGTAATCCTCGTTGTGGTCACGATTCCCGTGCGCTTGCCTTCCTTCTGGGCCCACTCCATGATAGTCTCAATCCGGCCCAGCT |
| droPer1 | super_7:3056347-3056483 + | GCACTCCCAGTCGCGATCGTAGATGTGGGCGTAGGTGGCCGCAGGCGTGGCATGCGTAATCCTCGTTGTGGTCACGATTCCCGTGCGCTTGCCTTCCTTCTGGGCCCACTCCATGATAGTCTCAATCCGGCCCAGCT |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
| dG=-33.3, p-value=0.009901 | dG=-33.0, p-value=0.009901 | dG=-32.7, p-value=0.009901 | dG=-32.5, p-value=0.009901 | dG=-32.4, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-34.6, p-value=0.009901 | dG=-34.6, p-value=0.009901 | dG=-34.4, p-value=0.009901 | dG=-34.4, p-value=0.009901 | dG=-34.4, p-value=0.009901 |
|---|---|---|---|---|
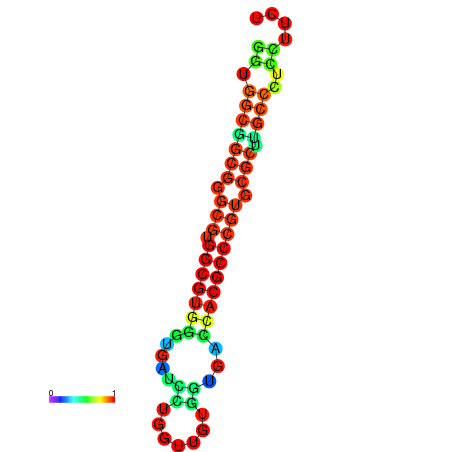 |
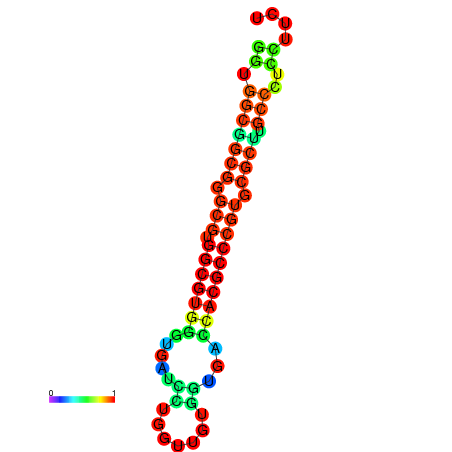 |
 |
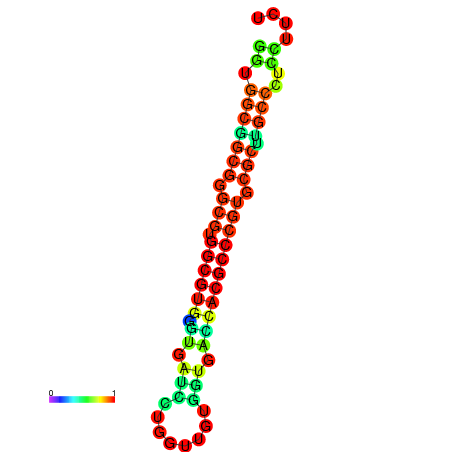 |
 |
| dG=-32.5, p-value=0.009901 | dG=-32.2, p-value=0.009901 | dG=-32.2, p-value=0.009901 | dG=-31.9, p-value=0.009901 |
|---|---|---|---|
 |
 |
 |
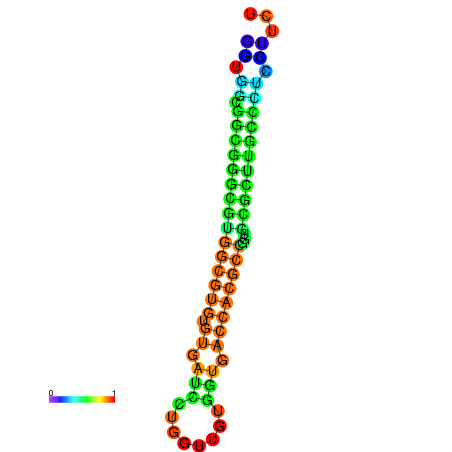 |
| dG=-26.4, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-25.9, p-value=0.009901 |
|---|---|---|---|---|
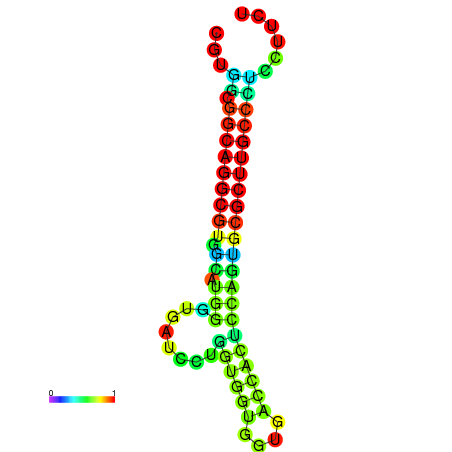 |
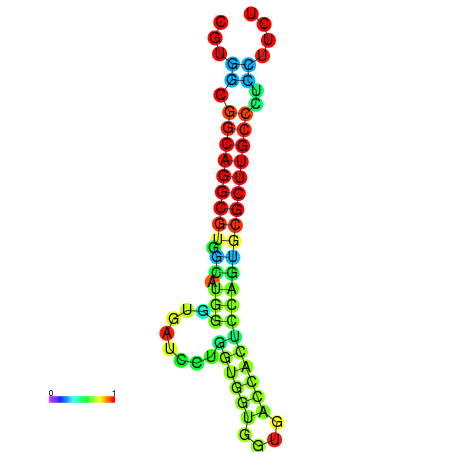 |
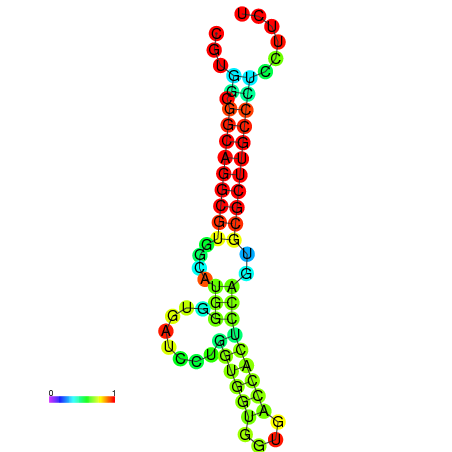 |
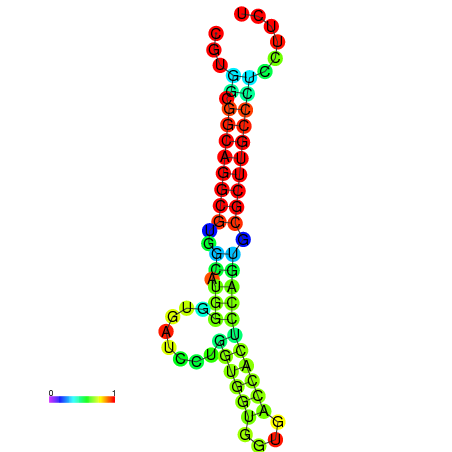 |
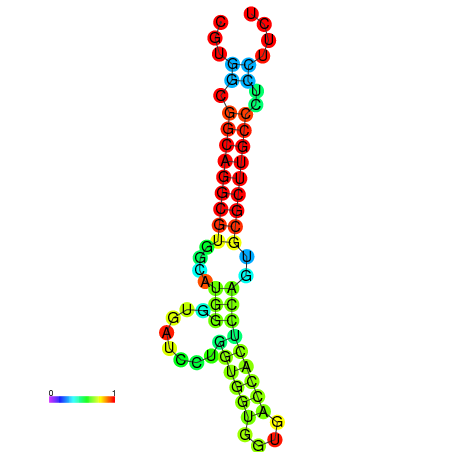 |
| dG=-26.6, p-value=0.009901 | dG=-26.4, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-26.1, p-value=0.009901 | dG=-26.0, p-value=0.009901 |
|---|---|---|---|---|
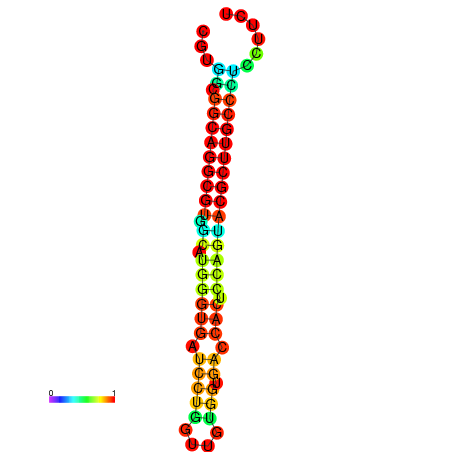 |
 |
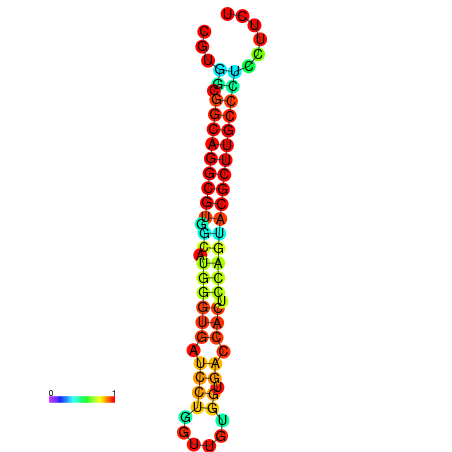 |
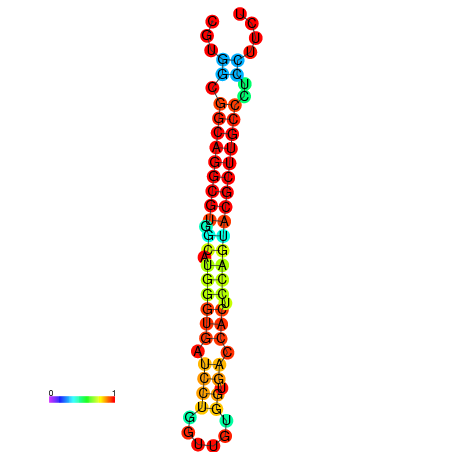 |
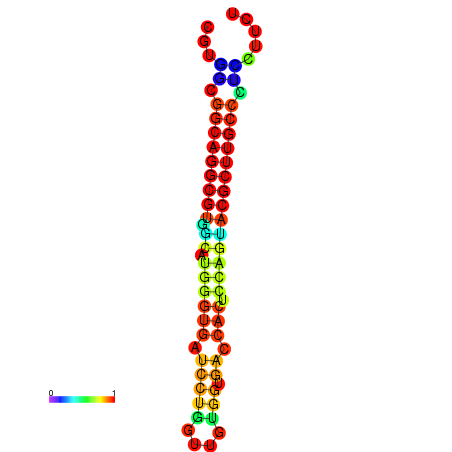 |
| dG=-16.4, p-value=0.009901 | dG=-16.3, p-value=0.009901 | dG=-15.7, p-value=0.009901 | dG=-15.4, p-value=0.009901 |
|---|---|---|---|
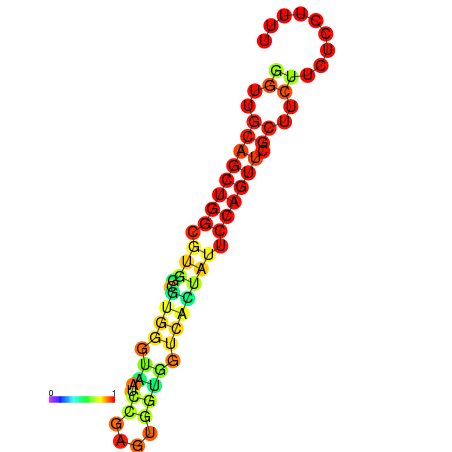 |
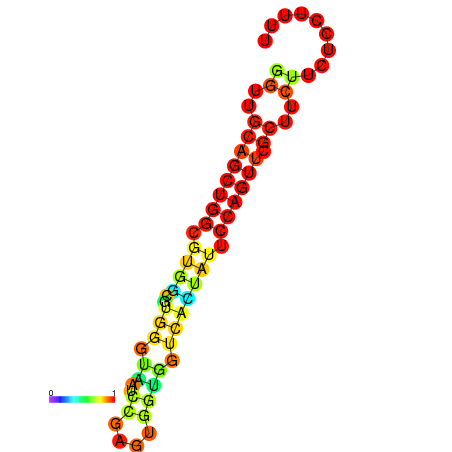 |
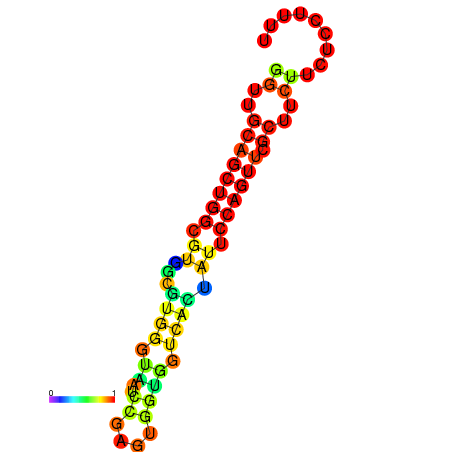 |
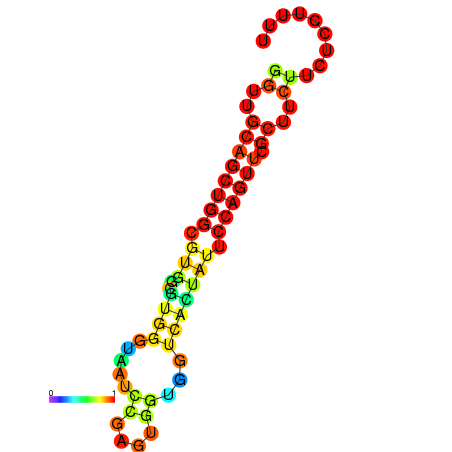 |
| dG=-20.5, p-value=0.009901 | dG=-20.0, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.6, p-value=0.009901 |
|---|---|---|---|---|
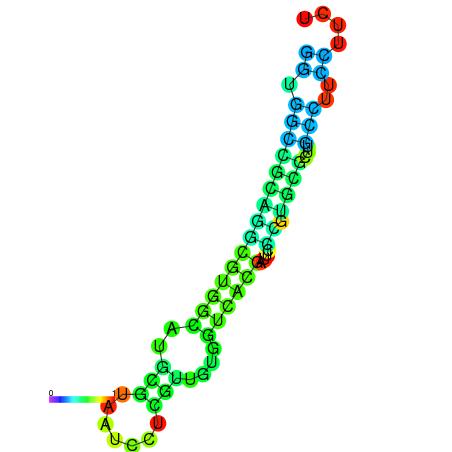 |
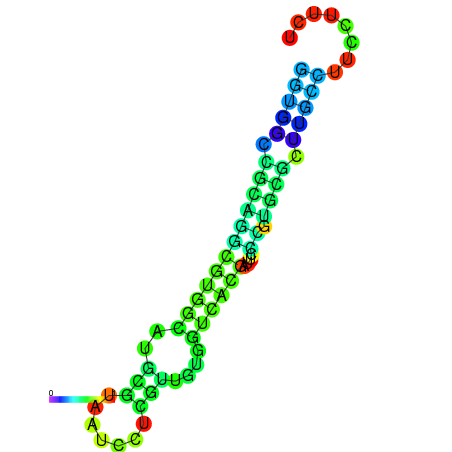 |
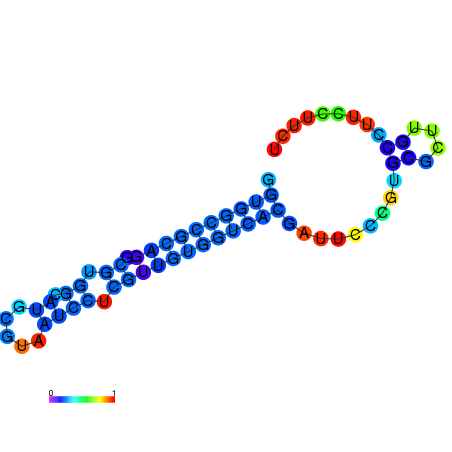 |
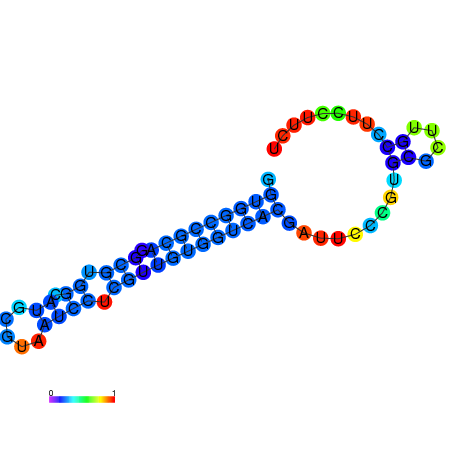 |
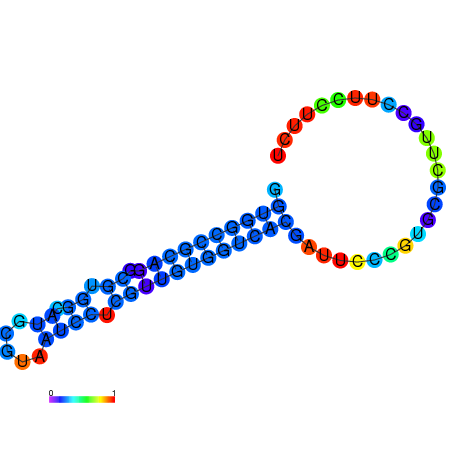 |
| dG=-20.5, p-value=0.009901 | dG=-20.0, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.9, p-value=0.009901 | dG=-19.6, p-value=0.009901 |
|---|---|---|---|---|
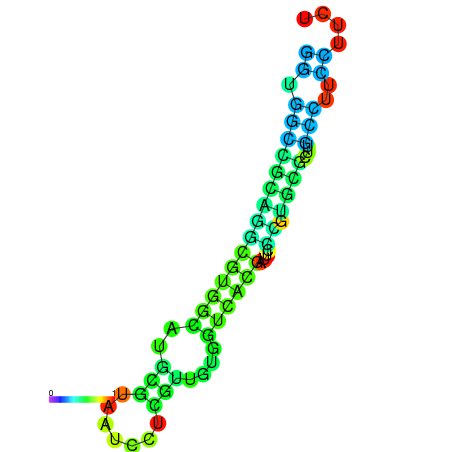 |
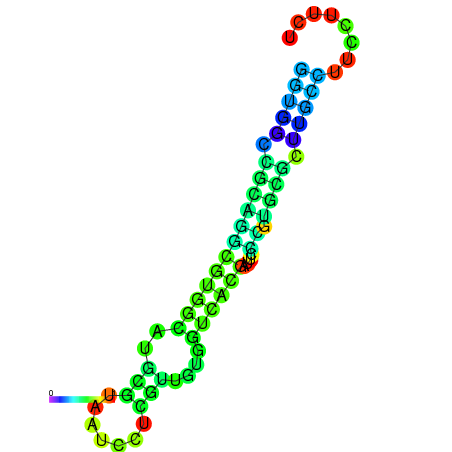 |
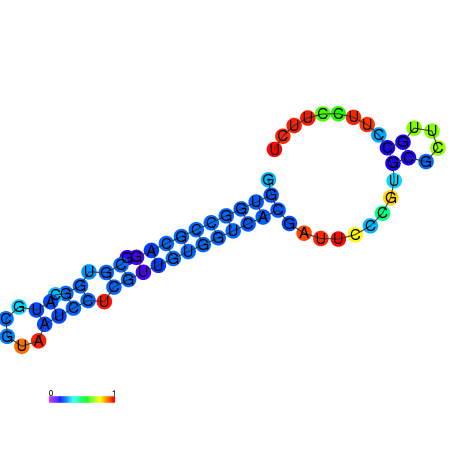 |
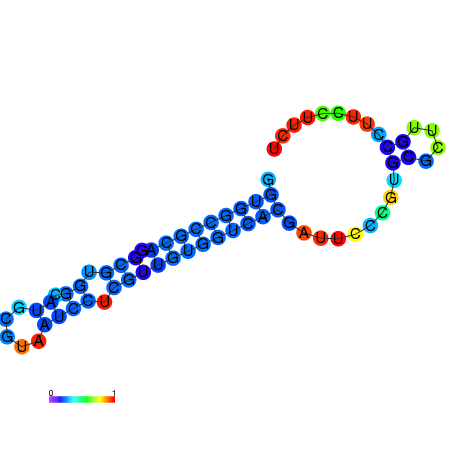 |
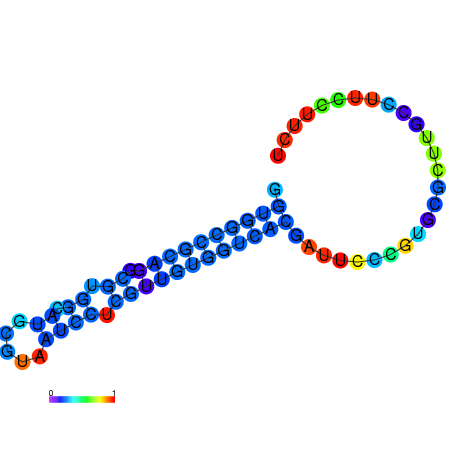 |
| dG=-11.1, p-value=0.009901 | dG=-11.0, p-value=0.009901 | dG=-10.9, p-value=0.009901 | dG=-10.7, p-value=0.009901 | dG=-10.7, p-value=0.009901 |
|---|---|---|---|---|
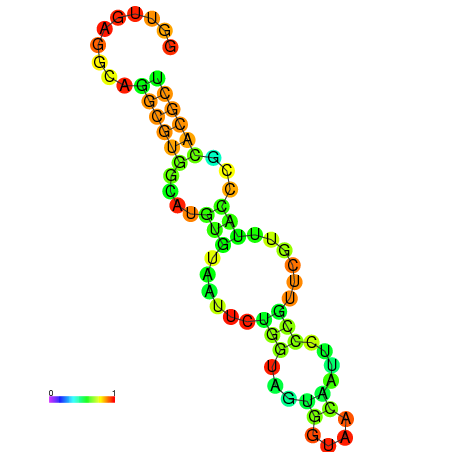 |
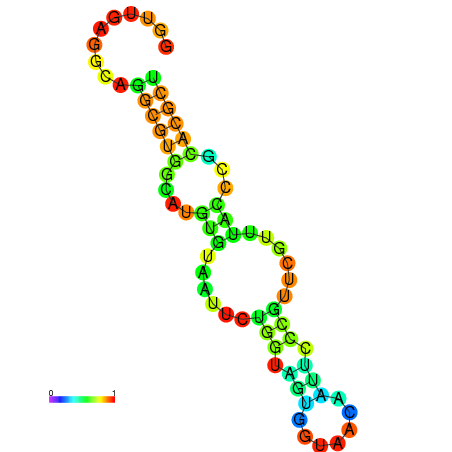 |
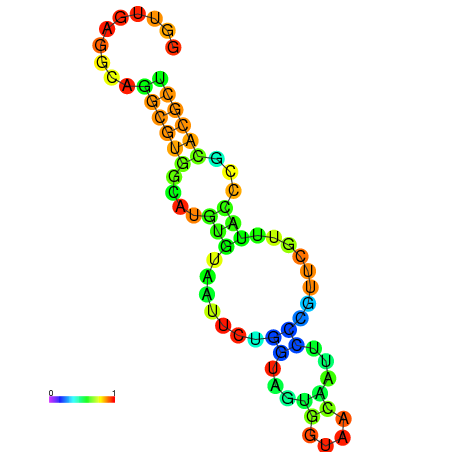 |
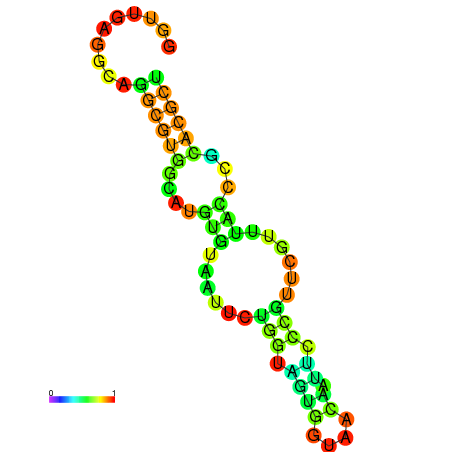 |
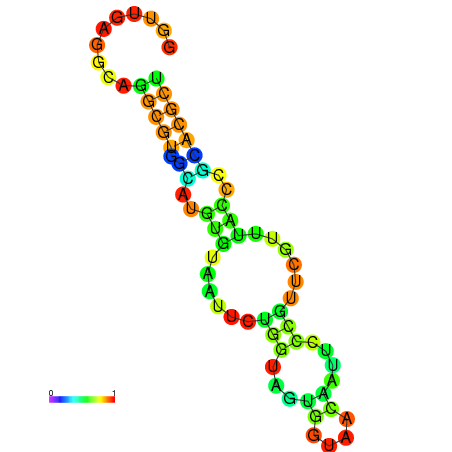 |
| dG=-29.1, p-value=0.009901 | dG=-28.8, p-value=0.009901 | dG=-28.5, p-value=0.009901 | dG=-28.4, p-value=0.009901 | dG=-28.2, p-value=0.009901 |
|---|---|---|---|---|
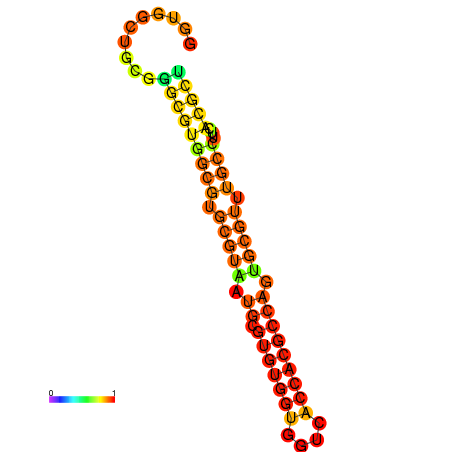 |
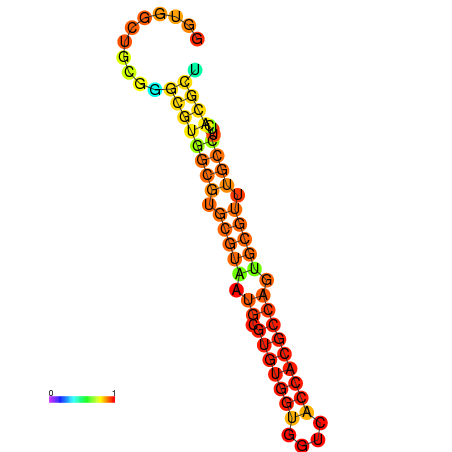 |
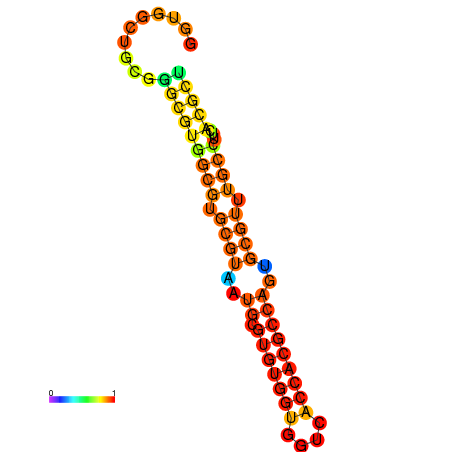 |
 |
 |
| dG=-26.7, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-25.7, p-value=0.009901 |
|---|---|---|
 |
 |
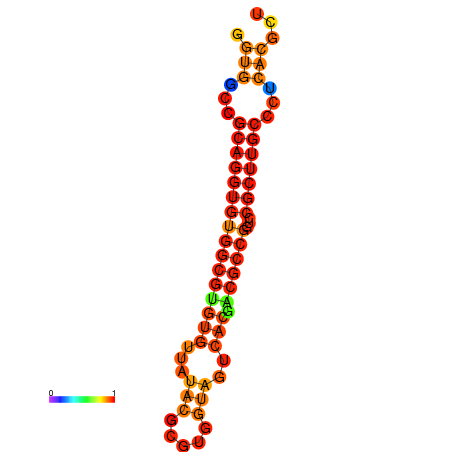 |
| dG=-21.7, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.4, p-value=0.009901 | dG=-21.3, p-value=0.009901 | dG=-21.2, p-value=0.009901 |
|---|---|---|---|---|
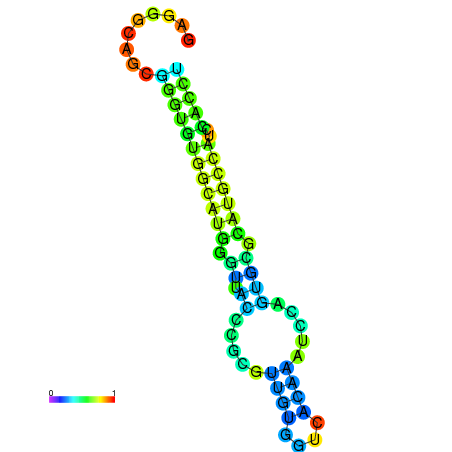 |
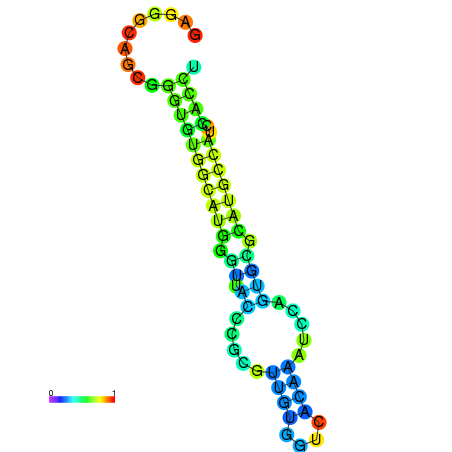 |
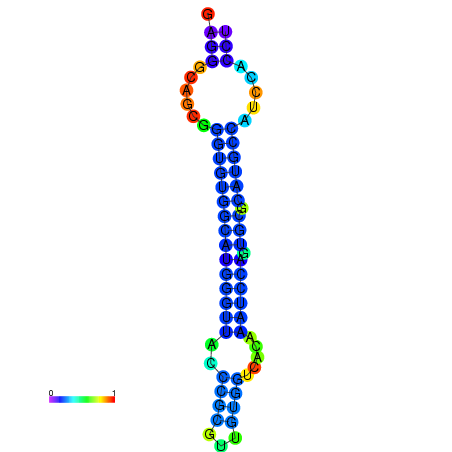 |
 |
 |
Generated: 03/07/2013 at 05:46 PM