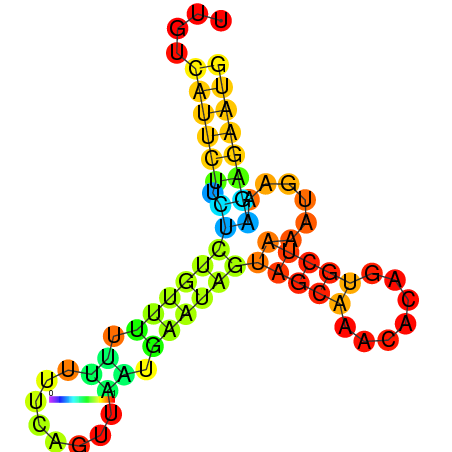
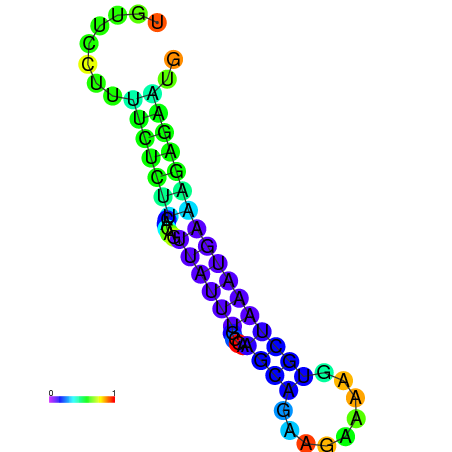
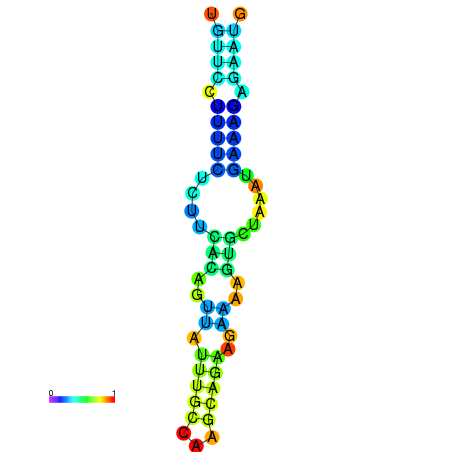
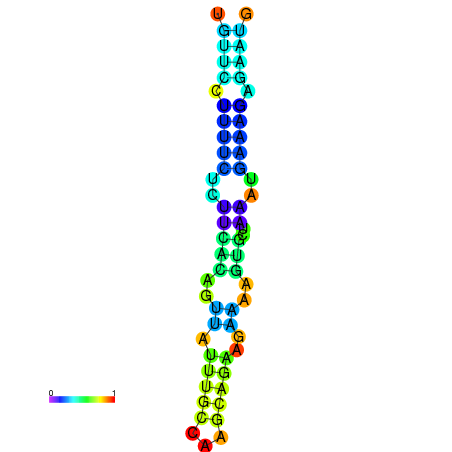
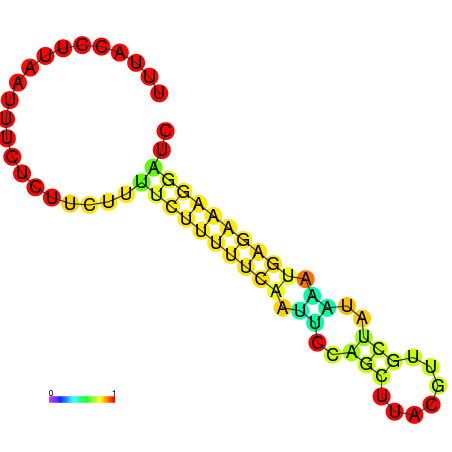
| dm3 |
chr2L:16528359-16528506 - |
ATTTTAAACAACTA-TTTCGG-------ATCGTTTG---------------AATGACCAATTTTGT-------------CTTTTATTTTCTCT--G--------------T-G-----TTTT-----CAGTTAATG-AAAGAT-AG-TGAACACAGTGCTAAATGAAAGATAATGAGACCCAGTCGACAGGACCC---------AAGAAGACGATGTTGATTG |
| droSim1 |
chr2L_random:601476-601603 - |
ATTT------------------------ATTT-TTG---------------AATGACCAATTTTGT-------------CTT---TCTTCTCT--G--------------T-C-----GTTT-----CAGTTAATG-AAAAGT-AG-TAAACACAGTGCTAAATGAAAGAGAATGAGACCCAGTCGACAGGACCC---------AAGAAGACGATGTTGATTG |
| droSec1 |
super_7:174891-175018 - |
ATTT------------------------ATTT-TTG---------------AATGACCAATTTTGT-------------CTT---TCTTCTCT--T--------------T-C-----TTTT-----CAGTTAATG-AAAAGT-AG-TAAACACAGTGCTAAATGAAAGAGAATGAGACCCAGTCGACAGGACCC---------AAGAAGACGATGTTGATTG |
| droYak2 |
chr2R:4574442-4574584 + |
ATTTTATAAAACTA-TGGGGG-------ATAC-TTA---------------GATAATTAATTTTGT-------------CAT---TGTTCTTT--G--------------T-C------TTT-----CAGTTAATG-AAAAGT-AG-TAAACACAGTGCTAAATGAAAGAGAATGAGACGCAGTCGACAGGACCC---------AAGAAGACGATGTTGATTG |
| droEre2 |
scaffold_4845:19592406-19592554 - |
ATTTTATGCAACTA-TTGTGT-----GTATCT-TTA---------------AATGACTAATTTTGT-------------CAT---TCTTCTCT--G------TTT-----T-T-----TTTT-----CAGTTAATG-AATAGT-AG-CAAACACAGTGCTAAATGAAAGAGAATGAGACGCAGTCGACAGGACCC---------AAGAAGACGATGTTGATTG |
| droAna3 |
scaffold_12916:16031637-16031788 + |
ATGTT------------------------TAA-ATG---------------ATTAAAAAATTATGTTCAACTAATTATTAA-------TTTAT--A------TTTT-ATTGTCTTCCCATTA-----CAGTTAATGAAAAAGT-AATTTAAAAAATCGCTAAATGAAAGAGAATGAGACCCAGTCGACATTTCCC---------AAGAAGACCATGTTGATTG |
| dp4 |
chr4_group3:8251613-8251715 - |
TT-----------------------------------------------------------------------------------TGTTCCTTTTC--------------T-C-----TTTA-----CAGTTATTTGCCAAGC-AG--AAGAAAAGTGCTAAATGAAAGAGAATGAGACACAGTCGACAGACGAA---------CAGAAGACGATGTTGATTG |
| droPer1 |
super_1:9700572-9700674 - |
TT-----------------------------------------------------------------------------------TGTTCCTTTTC--------------T-C-----TTCA-----CAGTTATTTGCCAAGC-AG--AAGAAAAGTGCTAAATGAAAGAGAATGAGACACAGTCGACAGACGAA---------CAGAAGACGATGTTGATTG |
| droWil1 |
scaffold_180772:4810613-4810759 - |
AAGTTTACCAATTAATAGCAA-------ATTG-TTGGTATTTCCATTTTAAAATTGCTAACATTTA---------CCTTAA-------TTTCT--C------TTCT-TTTC-T-----TTTT-----CAAT---------TCC-AG-CTTACGTTGCTATAAATGAGAAAGGATCAA------------GATCAA---------ACGAAAAGGATGTTGATTG |
| droVir3 |
scaffold_13052:1601321-1601473 - |
ATATTGCGCAACCA-TTCTGGTCCGAATTTCA-TTG---------------AGTCAGTCATTTTGG----GTAACTCTT------------GA--GTTTATTTCTTAATTGTC------TTTCGCTGAAATTGTGCAATAAAT-AA-CA---ATTT---------------TACCATCAGCATTAGCCAGCACAC---------ACACGCACGCTATCGGTTT |
| droMoj3 |
scaffold_6500:21042727-21042874 - |
AATTA------------------------TAA-TTG---------------AACACTTGA-TGCGATTGCCAAATTTTTGAT---TTTTGGTTCTA--------------T-T-----TTTA-----GAGTTACAG-CTAAAT-GA-AA---ACTATACCAATTATAAAATCAAAAGGGCTACACAAGAACATTTATGATTTCAAAGAAAAGTGTTTCATTTG |
| droGri2 |
scaffold_14822:853797-853916 - |
GCATG-----------------------------------------------ATGACTTGTGGTCT-------------CTA---TTCTATAT--G--------------C-T-----GGTT-----GAGTTTGTG-AATGGTCAG-TTGCCGTGGCGAAAGATAAAAGACAAG--GAACAGGTTGGATTGACCA---------AAGAGAAATATGATCATAG |