
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:16024273-16024406 + | CACGCTCACCAATCCCATGACCGGCGAGGAGATGCGATTCCTCGAGCTGCTCACCGCCTTTGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGAGTCGATATGCACACGCTCAACAACATGGAGCGCACCG |
| droSim1 | chr3L:15362441-15362574 + | CACGCTCACCAATCCCATGACCGGCGAGGAGATGCGCTTCCTCGAGCTGCTCACCGCCTTTGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGAGTCGATATGCACACGCTCAACAACATGGAGCGCACCG |
| droSec1 | super_0:8124906-8125039 + | CACGCTCACCAATCCCATGACCGGCGAGGAGATGCGCTTCCTCGAGCTGCTCACCGCCTTTGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGAGTCGATATGCACACGCTCAACAATATGGAGCGCACCG |
| droYak2 | chr3L_random:1624866-1624999 - | CACGCTCACGAATCCCATGACCGGCGAGGAGATGCGCTTCCTGGAGCTGCTCACCGCCTTCGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGCGTCGACATGCACACGCTCAACAACATGGAGCGCACCG |
| droEre2 | scaffold_4784:18269422-18269555 + | CACGCTCACGAATCCCATGACCGGCGAGGAGATGCGCTTCCTGGAGCTGCTCACCGCCTTCGGCTTGGACATCGAGGCGGTGGCCGCTGCACTGGGCGTCGACATGCACACGCTCAACAACATGGAGCGCACCG |
| droAna3 | scaffold_13337:16593401-16593534 - | CACCCTCACGAACCCCCTGACCGGCGAGGAGATGCGCTTTCTGGAGCTGCTCACGTCCTTCGGCATGGACATCGAGACAGTGGCCACCGCCCTGGGCGTGGATATGCACACGCTGAACAACATGGAGCGGACGG |
| dp4 | chrXR_group8:4268508-4268641 + | CACACTGACCAATCCGCTGACCGGCGAGGAGATGCGCTTCCTGGAGCTGCTCACCTCGTTCGGACTGGACATCGAGGCGGTGGCGGCGGCCCTGGGCGTCGACATGCACACCCTCAACAACATGGAGCGGACGG |
| droPer1 | super_29:501948-502081 + | CACACTGACCAATCCGCTGACCGGCGAGGAGATGCGCTTCCTGGAGCTGCTCACCTCGTTCGGGCTGGACATCGAGGCGGTGGCGGCGGCCCTGGGCGTCGACATGCACACCCTCAACAACATGGAGCGGACGG |
| droWil1 | scaffold_181024:787133-787266 + | CACATTAACCAATCCACTGACAGGTGAAGAGATGCGTTTCCTTGAGCTGCTCACCTCCTTCGGTTTGGATATTGAGGCAGTGGCAGCGGCGCTTGGCGTCGATATGCATACCCTCAACAATATGGAACGTACCG |
| droVir3 | scaffold_13049:9209848-9209981 + | CACGCTAACAAATCCGCTGACCGGCGAGGAGATGCGCTTTCTCGAGCTGCTCACCTCCTTCGGCCTGGACATCGAGGCCGTCGCCGCCGCACTCGGCGTCGATATGCACACCCTGAACAACATGGAACGCACCG |
| droMoj3 | scaffold_6680:6136665-6136798 - | CACGCTCACGAATCCGCTGACCGGCGAGGAGATGCGCTTCCTCGAACTGCTCACCTCCTTCGGTCTGGACATCGAAGCCGTGGCCGCAGCACTCGGCGTCGATATGCACACGCTGAACAACATGGAACGAACGG |
| droGri2 | scaffold_15110:4981116-4981249 + | CACGCTCACGAATCCGCTAACCGGCGAGGAGATGCGCTTCCTCGAGCTGCTCACCTCATTCGGCTTGGACATTGAGGCGGTGGCAGCAGCGCTGGGTGTCGATATGCACACCCTGAATAACATGGAGCGAACGG |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3L:16024273-16024406 + | CACGCTCACCAATCCCATGACCGGCGAGGAGATGCGATTCCTCGAGCTGCTCACCGCCTTTGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGAGTCGATATGCACACGCTCAACAACATGGAGCGCACCG |
| droSim1 | chr3L:15362441-15362574 + | CACGCTCACCAATCCCATGACCGGCGAGGAGATGCGCTTCCTCGAGCTGCTCACCGCCTTTGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGAGTCGATATGCACACGCTCAACAACATGGAGCGCACCG |
| droSec1 | super_0:8124906-8125039 + | CACGCTCACCAATCCCATGACCGGCGAGGAGATGCGCTTCCTCGAGCTGCTCACCGCCTTTGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGAGTCGATATGCACACGCTCAACAATATGGAGCGCACCG |
| droYak2 | chr3L_random:1624866-1624999 - | CACGCTCACGAATCCCATGACCGGCGAGGAGATGCGCTTCCTGGAGCTGCTCACCGCCTTCGGCTTGGACATCGAGGCGGTGGCCGCCGCTCTGGGCGTCGACATGCACACGCTCAACAACATGGAGCGCACCG |
| droEre2 | scaffold_4784:18269422-18269555 + | CACGCTCACGAATCCCATGACCGGCGAGGAGATGCGCTTCCTGGAGCTGCTCACCGCCTTCGGCTTGGACATCGAGGCGGTGGCCGCTGCACTGGGCGTCGACATGCACACGCTCAACAACATGGAGCGCACCG |
| Species | Read pileup | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||
| dp4 |
|
| dG=-39.0, p-value=0.009901 | dG=-38.5, p-value=0.009901 |
|---|---|
 |
 |
| dG=-34.3, p-value=0.009901 | dG=-34.0, p-value=0.009901 | dG=-33.8, p-value=0.009901 | dG=-33.5, p-value=0.009901 | dG=-33.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-34.3, p-value=0.009901 | dG=-34.0, p-value=0.009901 | dG=-33.8, p-value=0.009901 | dG=-33.5, p-value=0.009901 | dG=-33.3, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-34.2, p-value=0.009901 | dG=-34.2, p-value=0.009901 | dG=-33.7, p-value=0.009901 | dG=-33.7, p-value=0.009901 | dG=-33.5, p-value=0.009901 |
|---|---|---|---|---|
 |
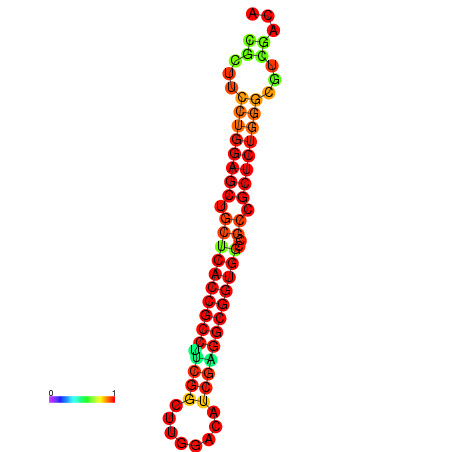 |
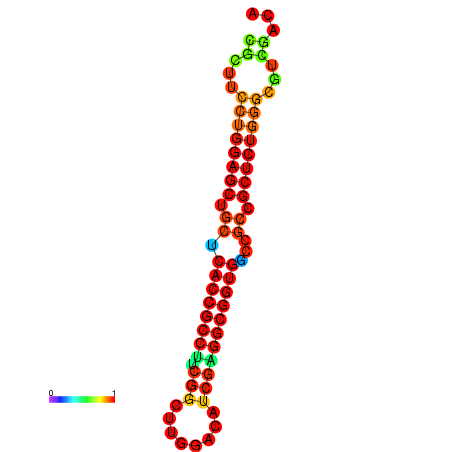 |
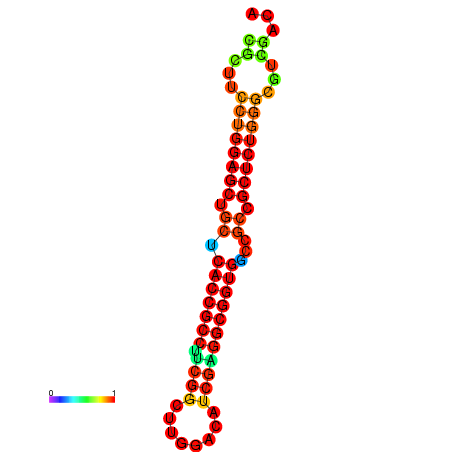 |
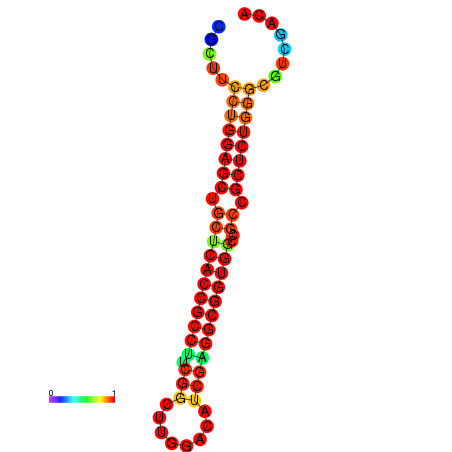 |
| dG=-34.3, p-value=0.009901 | dG=-34.3, p-value=0.009901 |
|---|---|
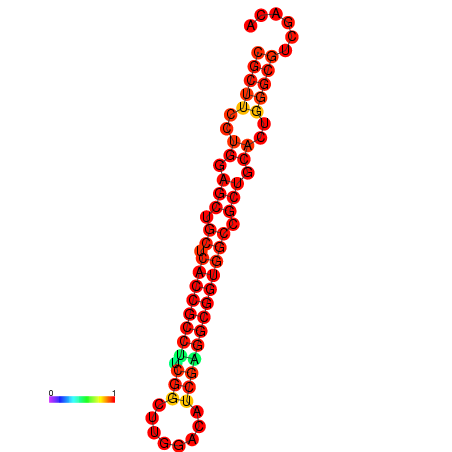 |
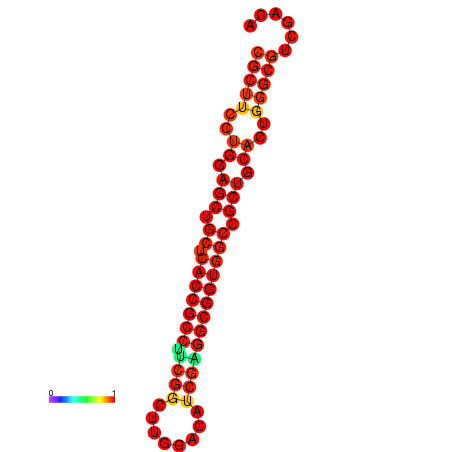 |
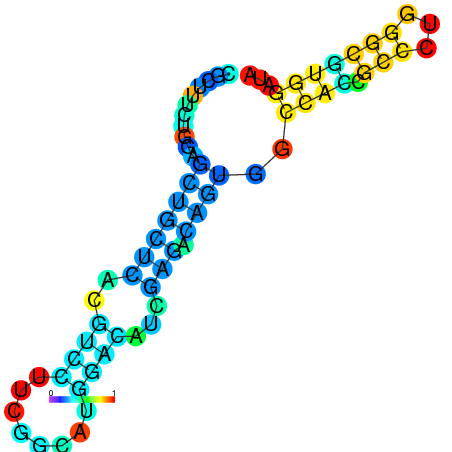
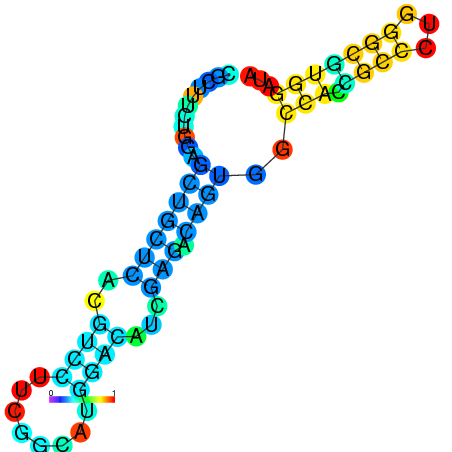
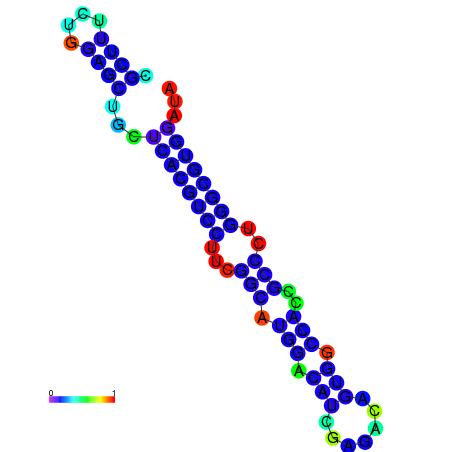
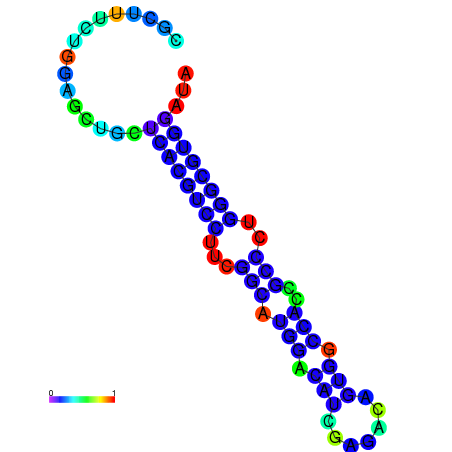
| dG=-22.0, p-value=0.009901 | dG=-22.0, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.9, p-value=0.009901 | dG=-21.6, p-value=0.009901 | dG=-19.7, p-value=0.009901 |
|---|---|---|---|---|---|
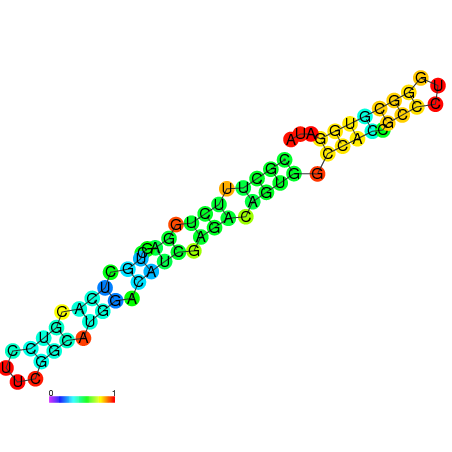 |
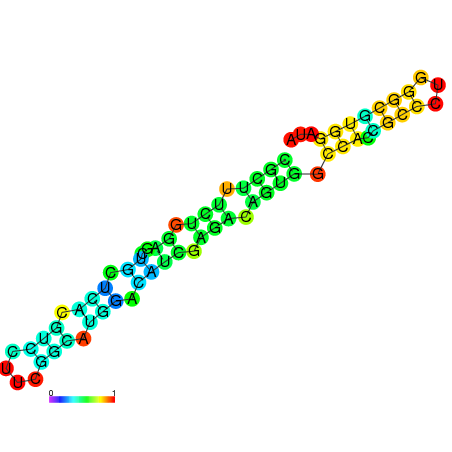 |
 |
 |
 |
 |
| dG=-25.2, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.6, p-value=0.009901 |
|---|---|---|---|---|
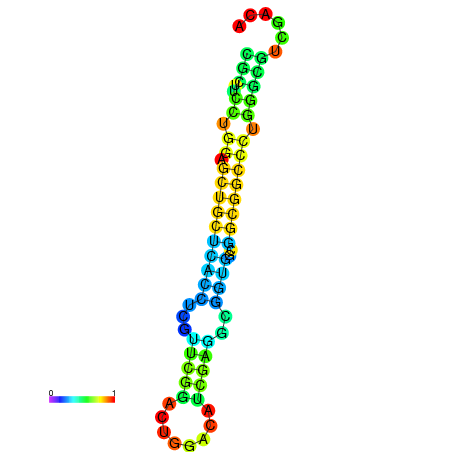 |
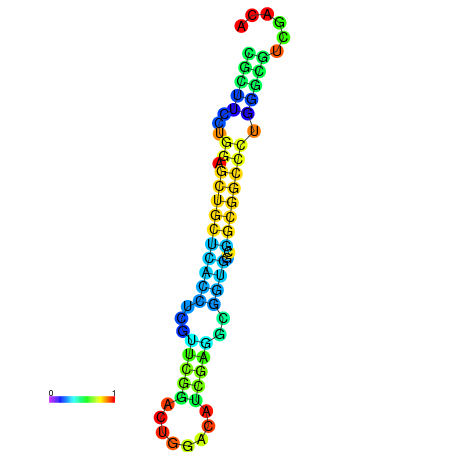 |
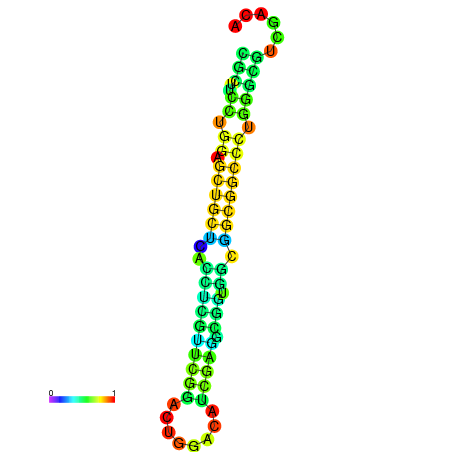 |
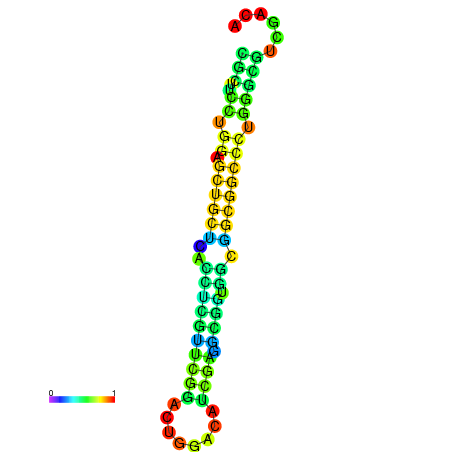 |
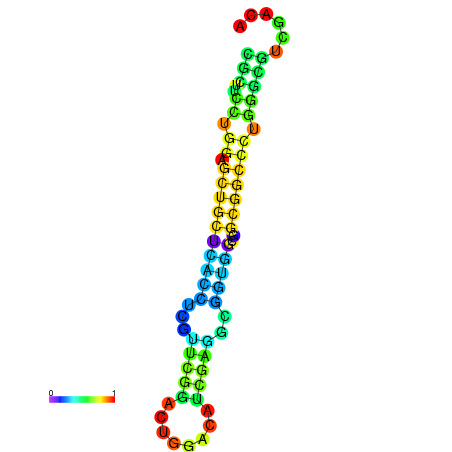 |
| dG=-26.4, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
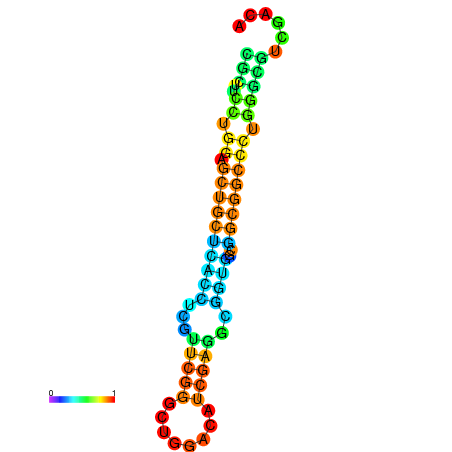 |
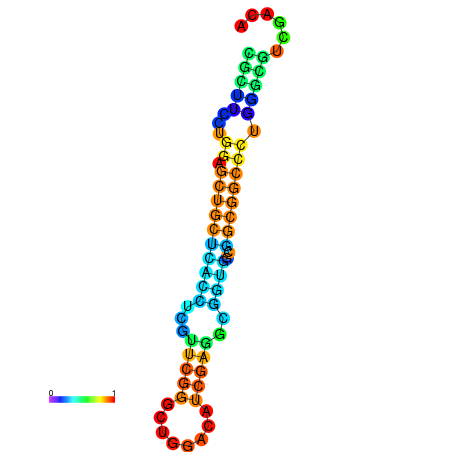 |
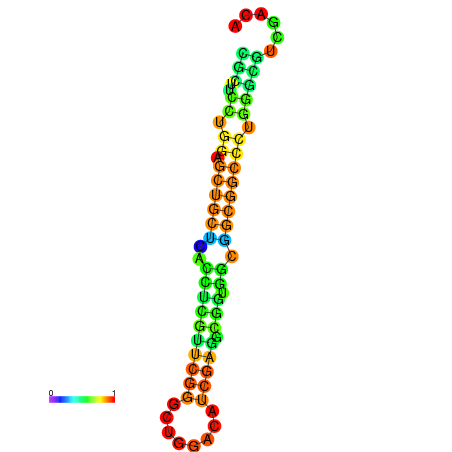 |
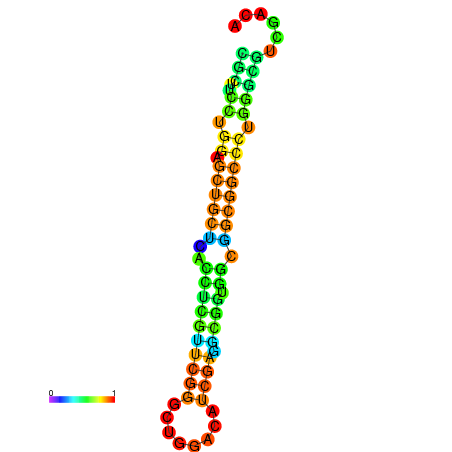 |
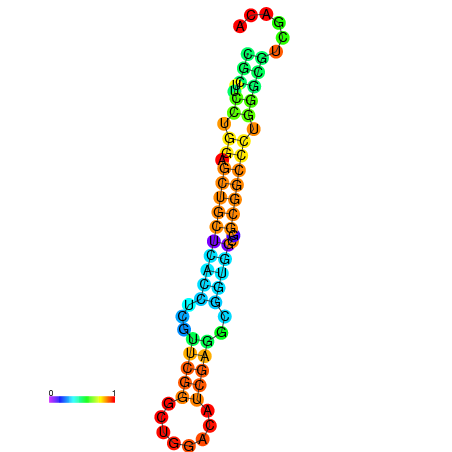 |
| dG=-23.3, p-value=0.009901 | dG=-23.3, p-value=0.009901 | dG=-22.9, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-22.8, p-value=0.009901 | dG=-21.1, p-value=0.009901 |
|---|---|---|---|---|---|
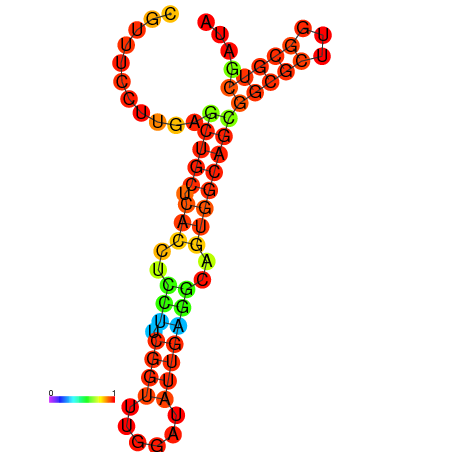 |
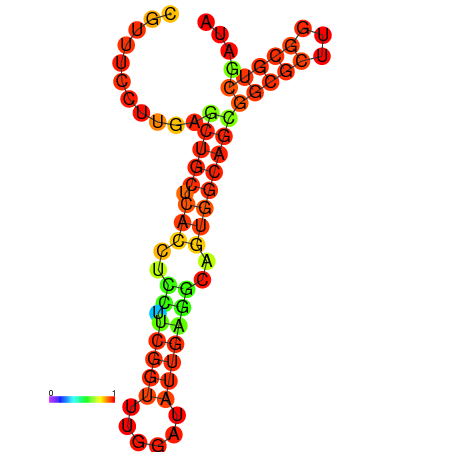 |
 |
 |
 |
 |
| dG=-24.5, p-value=0.009901 | dG=-23.5, p-value=0.009901 | dG=-20.6, p-value=0.009901 |
|---|---|---|
 |
 |
 |
| dG=-17.7, p-value=0.009901 | dG=-17.2, p-value=0.009901 | dG=-16.9, p-value=0.009901 | dG=-16.7, p-value=0.009901 |
|---|---|---|---|
 |
 |
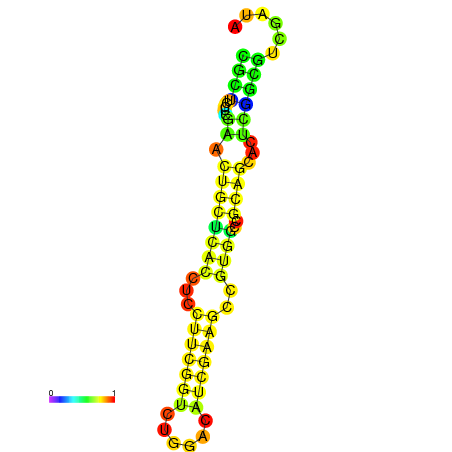 |
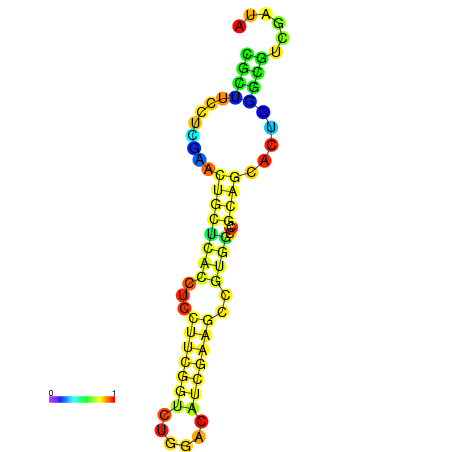 |
| dG=-22.0, p-value=0.009901 | dG=-21.5, p-value=0.009901 | dG=-21.3, p-value=0.009901 | dG=-21.2, p-value=0.009901 | dG=-21.1, p-value=0.009901 |
|---|---|---|---|---|
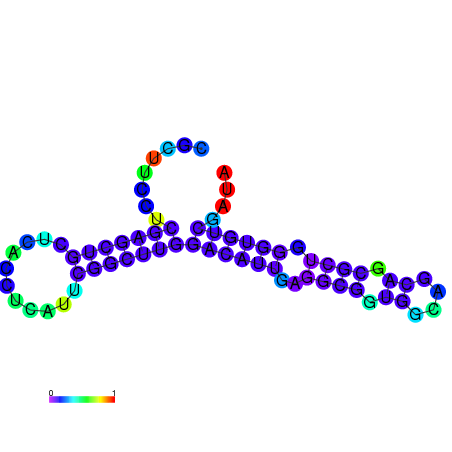 |
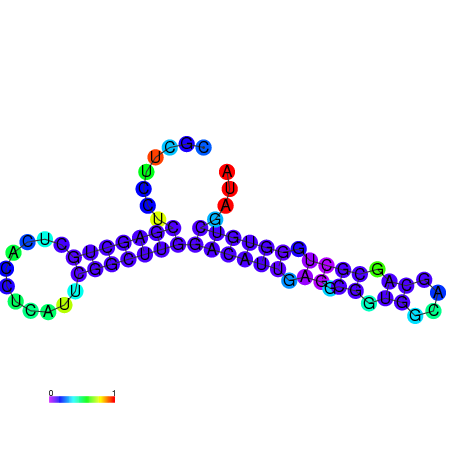 |
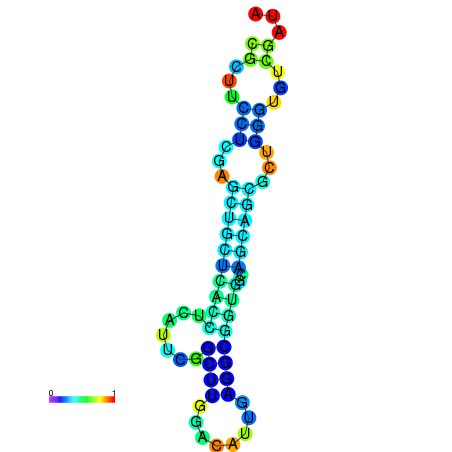 |
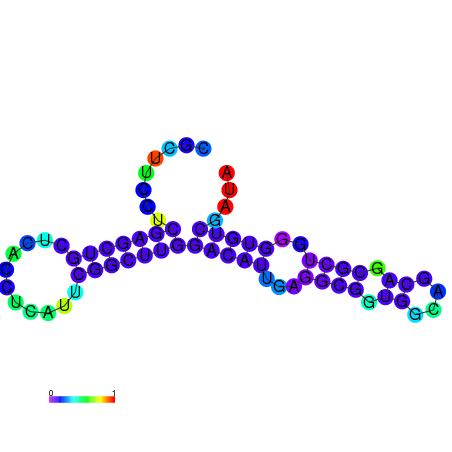 |
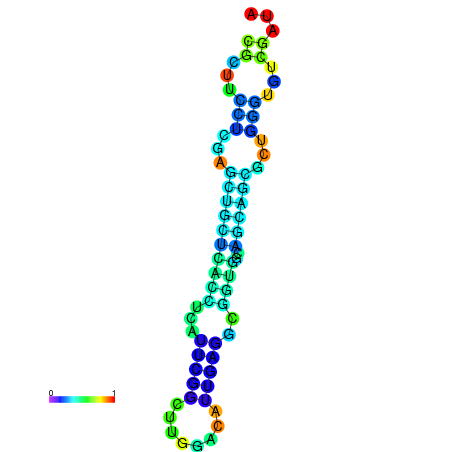 |
Generated: 03/07/2013 at 05:36 PM