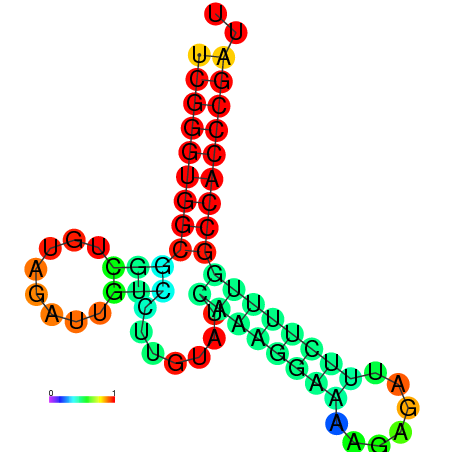
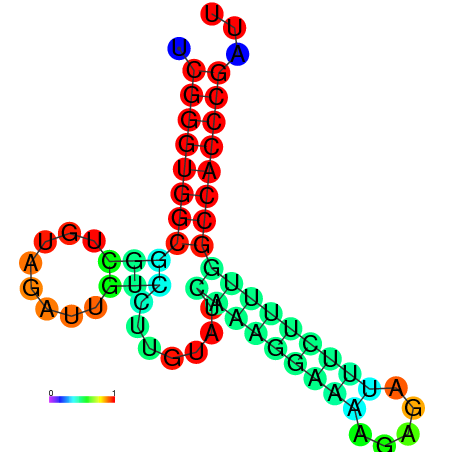
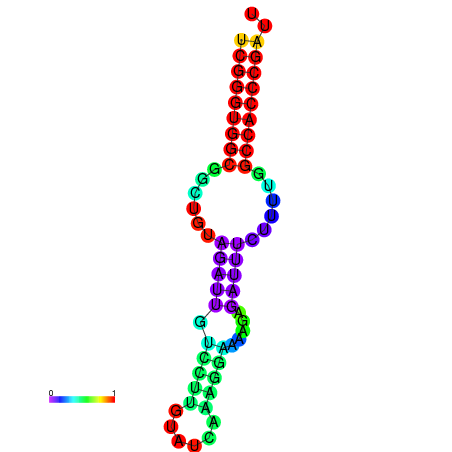
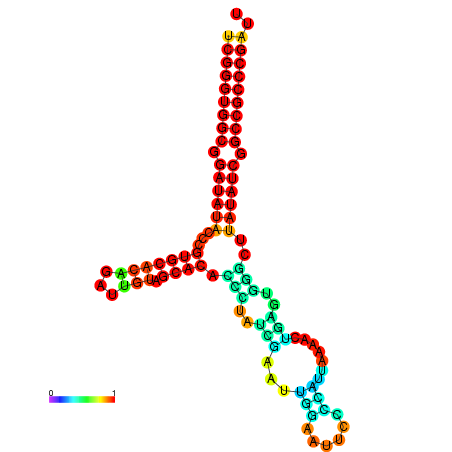
| dm3 |
chr3L:9137677-9137812 + |
CCAGACTCAGACGGACTTA----TCGCAA----CTTATCGAT------CGGGTGGCGGATATA----------GATCA-----------------TTGGTT---------------------------------------------------------------------------------------------------------------ATCCCTTAATTAAAGC--------AAACAAGATTTGTTTCGGCCGCCC--GATTTTACATACAACGAC-----------------ACGAATAGCAGG-----------------------------ATAT-AG |
| droSim1 |
chr3L:8545595-8545743 + |
CCTGACTCAGACGGACTTA----TCGCAG----CTTATCGAT------CGGGTGGCGGATATA----------GATCATAGCAAGCCCTATCGAATTGGTT---------------------------------------------------------------------------------------------------------------ATACCT--ATTAAAGC---------AACTAGGTTTGTTTCGGCCGCCC--GATTTTACATACAACGAC-----------------ACAA-TAGCAGG-----------------------------ATAT-AG |
| droSec1 |
super_0:1396669-1396817 + |
CCTGACTCAGACGGACTTA----TCGCAG----CTTATCGAT------CGGGTGGCGGATATA----------GATCATAGCAACCCCTATCGAATCGGTT---------------------------------------------------------------------------------------------------------------ATACCT--ATTAATGC---------AACTAGGTTTGTTTCGGCCGCCC--GATTTAACATACAACGAC-----------------ACAA-TAGCAGG-----------------------------ATAT-AG |
| droYak2 |
chr3L:2783249-2783373 - |
CCAGACTCAGACGGACTA-------GCAA----CTTCGCGAT------CGGGTGGCGGCTGTA----------GATT-------------------------------------------------------------------------------------------------------------GT-----------------CCTTGT---------ATCAAAGG--------AAAAGAGATTTCTTTTGGCCACCC--GATTCAAAATACAACGAC-----------------ACAA-TAGCAGG-----------------------------ATAT-AG |
| droEre2 |
scaffold_4784:4908876-4909031 - |
CCTGACTCAGACGGACTA-------GCAA----CTTATCGAT------CGGGTGGCGGATATACCCGTGCACAGATTGTAGCACACCCTATCGAATTGGAA---------------------------------------------------------------------------------------------------------------TTCCCC--ATTAAAAC--------TGAGTGGGCTTATATCGGCCGCCC--GATT-CACATACAACGAC-----------------ACAA-TAGCAGG-----------------------------ATAT-AG |
| droAna3 |
scaffold_13337:19535893-19536018 + |
CACTGCGATCTCGAGTACT----TT-GATCTCTCTTCTCAGGGCCCC-AAAAAGTATGCTAT-------------------------------------------------------------------------------------------------------------GA-------AAT-ATTTCGTTTGC--------------------------TAAAGTG--------TTTCAAGGATTCTTTTGCGAGCCC--ACCT------------CA-----------------AGGA-TGTCAAA-----------------------------CTCT-AG |
| dp4 |
chrXR_group5:393309-393508 - |
C-AGACTTAGACGGACTTCAATCTAGCAG----TTTAACGAT------CGGGAAACGGGAATA----------AACCAAAGT---------------------------------------------------CGGCAGCCTATCCATCAAAATTCCAATATAATCACAACAGAAA-----TC-CAATTTGTTGC----------------------------------------------TGGTTTATTTACGTTTCTC--GAGC-CACATACAACGAC-----------------ACAA-CAGCAGGTCTTCAGATAACTCATATGAAATATACTTATGC-GG |
| droPer1 |
super_9:3484001-3484112 - |
GCATACATAGGCATCGCA----------T----CC---------------------------------------------------------------------------------------------------------------ATCGGGGTCAC---------CCGGTGGAGGATAAA-TTTTGC-----------------GGTGGTAACCCCGCAACACAAGC--------A--------------------------CTCACACACACAGCGAC-----------------CCAA-ACACGTG-----------------------------CAATAAC |
| droWil1 |
scaffold_181096:6035694-6035827 - |
TCAGGCCTAAAATTAGTTG----CCATAA----CT-CTCATT------TATATAGTAGATTTG----------ATGCTTTTT-----------------------------------------------TGTT----------------------------------------------------------TTTT---------------------------TAATAT---------ATTAAAATTTTTATTA-------------ATGCATTTCA--GTGTAAAAATTAAAATGACACATTTTTCCTT-----------------------------ACGT-AA |
| droVir3 |
scaffold_13246:101577-101715 - |
CAGG---CAGACGAAAGTA----TCTC-----------CGAC--CC--CATGAAGTGTATATA------------TTC-----------------TTGATCAGCATCAATAGCTGAGTCGATGTAGCCATGTCCGTCTGCC----------------------------------------------------------------------------------------------------------TGTCCGTCCGTCC--GTCCGTATGTATGA--ACGCA--------------AGGA-TCTCAGA-----------------------------ACCT-TT |
| droMoj3 |
scaffold_6540:2381798-2381917 - |
GTAGATTCAGTTGTACTTT----ATAGAA----TT--TCGAT-----TTA---------------------------------------------------------------------------------------------------------------------TGTTAA-------AAT-TCATCCCTTAC---------------------------CAATAT---------ATATGTGTT-TTTTCAAATGCATGCATTT-TAATGGTATCAAC-----------------ACAATTTGAAAG-----------------------------TCTC-AT |
| droGri2 |
scaffold_15074:3698733-3698860 + |
CTGAACATAACTGAATAAA----TT--------------------------------------------------------------------------------------------------------------------------------------------CACAACAT-------ATT-TAGTCAGTTACTTATGTATATACT----------TGATACAATTGTGCAAACATGCAAAATTCCAATTAAATGTCA--CAATATATAAAAACTG----------------------A-TGCGAGA-----------------------------AGAT-TG |